Differential transcriptional invasion signatures from patient derived organoid models define a functional prognostic tool for head and neck cancer
- PMID: 38942893
- PMCID: PMC11315671
- DOI: 10.1038/s41388-024-03091-4
Differential transcriptional invasion signatures from patient derived organoid models define a functional prognostic tool for head and neck cancer
Abstract
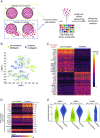
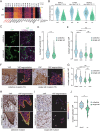
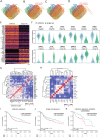
Clinical outcome for patients suffering from HPV-negative head and neck squamous cell carcinoma (HNSCC) remains poor. This is mostly due to highly invasive tumors that cause loco-regional relapses after initial therapeutic intervention and metastatic outgrowth. The molecular pathways governing the detrimental invasive growth modes in HNSCC remain however understudied. Here, we have established HNSCC patient derived organoid (PDO) models that recapitulate 3-dimensional invasion in vitro. Single cell mRNA sequencing was applied to study the differences between non-invasive and invasive conditions, and in a collective versus single cell invading PDO model. Differential expression analysis under invasive conditions in Collagen gels reveals an overall upregulation of a YAP-centered transcriptional program, irrespective of the invasion mode. However, we find that collectively invading HNSCC PDO cells show elevated levels of YAP transcription targets when compared to single cell invasion. Also, collectively invading cells are characterized by increased nuclear translocation of YAP within the invasive strands, which coincides with Collagen-I matrix alignment at the invasive front. Using gene set enrichment analysis, we identify immune cell-like migratory pathways in the single cell invading HNSCC PDO, while collective invasion is characterized by overt upregulation of adhesion and migratory pathways. Lastly, based on clinical head and neck cancer cohorts, we demonstrate that the identified collective invasion signature provides a candidate prognostic platform for survival in HNSCC. By uncoupling collective and single cell invasive programs, we have established invasion signatures that may guide new therapeutic options.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
PDLIM3 Regulates Migration and Invasion of Head and Neck Squamous Cell Carcinoma via YAP-Mediated Epithelial-Mesenchymal Transition.Int J Mol Sci. 2025 Mar 28;26(7):3147. doi: 10.3390/ijms26073147. Int J Mol Sci. 2025. PMID: 40243891 Free PMC article.
-
Catulin Based Reporter System to Track and Characterize the Population of Invasive Cancer Cells in the Head and Neck Squamous Cell Carcinoma.Int J Mol Sci. 2021 Dec 23;23(1):140. doi: 10.3390/ijms23010140. Int J Mol Sci. 2021. PMID: 35008571 Free PMC article.
-
Phenotype remodelling of HNSCC cells in the muscle invasion environment.J Transl Med. 2024 Oct 7;22(1):909. doi: 10.1186/s12967-024-05607-8. J Transl Med. 2024. PMID: 39375763 Free PMC article.
-
Patient-derived three-dimensional culture techniques model tumor heterogeneity in head and neck cancer.Oral Oncol. 2023 Mar;138:106330. doi: 10.1016/j.oraloncology.2023.106330. Epub 2023 Feb 9. Oral Oncol. 2023. PMID: 36773387 Free PMC article. Review.
-
The potential of hydrogel-free tumoroids in head and neck squamous cell carcinoma.Cancer Med. 2024 Aug;13(16):e70129. doi: 10.1002/cam4.70129. Cancer Med. 2024. PMID: 39169896 Free PMC article. Review.
Cited by
-
Head and neck tumor organoid grown under simplified media conditions model tumor biology and chemoradiation responses.Sci Rep. 2025 Jul 7;15(1):24221. doi: 10.1038/s41598-025-88082-5. Sci Rep. 2025. PMID: 40624315 Free PMC article.
-
A Multi-Omics Framework for Survival Mediation Analysis of High-Dimensional Proteogenomic Data.ArXiv [Preprint]. 2025 Mar 11:arXiv:2503.08606v1. ArXiv. 2025. PMID: 40160448 Free PMC article. Preprint.
References
-
- Cancer IC for OR (ICOR) in H and N, Ebrahimi A, Gil Z, Amit M, Yen TC, Liao CT, et al. Primary tumor staging for oral cancer and a proposed modification incorporating depth of invasion: an international multicenter retrospective study. Jama Otolaryngol Head Neck Surg. 2014;140:1138–48. 10.1001/jamaoto.2014.1548 - DOI - PubMed
-
- Bryne M, Boysen M, Alfsen CG, Abeler VM, Sudbø J, Nesland JM, et al. The invasive front of carcinomas. The most important area for tumour prognosis? Anticancer Res. 1998;18:4757–64. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

