Living in mangroves: a syntrophic scenario unveiling a resourceful microbiome
- PMID: 38943070
- PMCID: PMC11212195
- DOI: 10.1186/s12866-024-03390-6
Living in mangroves: a syntrophic scenario unveiling a resourceful microbiome
Abstract
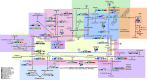
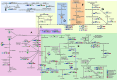
Background: Mangroves are complex and dynamic coastal ecosystems under frequent fluctuations in physicochemical conditions related to the tidal regime. The frequent variation in organic matter concentration, nutrients, and oxygen availability, among other factors, drives the microbial community composition, favoring syntrophic populations harboring a rich and diverse, stress-driven metabolism. Mangroves are known for their carbon sequestration capability, and their complex and integrated metabolic activity is essential to global biogeochemical cycling. Here, we present a metabolic reconstruction based on the genomic functional capability and flux profile between sympatric MAGs co-assembled from a tropical restored mangrove.
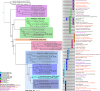
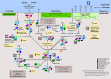
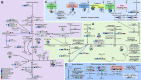
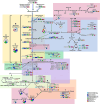
Results: Eleven MAGs were assigned to six Bacteria phyla, all distantly related to the available reference genomes. The metabolic reconstruction showed several potential coupling points and shortcuts between complementary routes and predicted syntrophic interactions. Two metabolic scenarios were drawn: a heterotrophic scenario with plenty of carbon sources and an autotrophic scenario with limited carbon sources or under inhibitory conditions. The sulfur cycle was dominant over methane and the major pathways identified were acetate oxidation coupled to sulfate reduction, heterotrophic acetogenesis coupled to carbohydrate catabolism, ethanol production and carbon fixation. Interestingly, several gene sets and metabolic routes similar to those described for wastewater and organic effluent treatment processes were identified.
Conclusion: The mangrove microbial community metabolic reconstruction reflected the flexibility required to survive in fluctuating environments as the microhabitats created by the tidal regime in mangrove sediments. The metabolic components related to wastewater and organic effluent treatment processes identified strongly suggest that mangrove microbial communities could represent a resourceful microbial model for biotechnological applications that occur naturally in the environment.
Keywords: Coastal sediment; MAG; Metabolic reconstruction; Metagenomics; Microbial metabolism.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Chambers LG, Guevara R, Boyer JN, Troxler TG, Davis SE. Effects of Salinity and Inundation on Microbial Community structure and function in a Mangrove Peat Soil. Wetlands. 2016;36:361–71. doi: 10.1007/s13157-016-0745-8. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

