Inhibition of BACE1 affected both its Aβ producing and degrading activities and increased Aβ42 and Aβ40 levels at high-level BACE1 expression
- PMID: 38944120
- PMCID: PMC11324814
- DOI: 10.1016/j.jbc.2024.107510
Inhibition of BACE1 affected both its Aβ producing and degrading activities and increased Aβ42 and Aβ40 levels at high-level BACE1 expression
Abstract
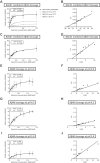
The beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) is the predominant β-secretase, cleaving the amyloid precursor protein (APP) via the amyloidogenic pathway. In addition, BACE1 as an amyloid degrading enzyme (ADE), cleaves Aβ to produce the C-terminally truncated non-toxic Aβ fragment Aβ34 which is an indicator of amyloid clearance. Here, we analyzed the effects of BACE1 inhibitors on its opposing enzymatic functions, i.e., amyloidogenic (Aβ producing) and amyloidolytic (Aβ degrading) activities, using cell culture models with varying BACE1/APP ratios. Under high-level BACE1 expression, low-dose inhibition unexpectedly yielded a two-fold increase in Aβ42 and Aβ40 levels. The concomitant decrease in Aβ34 and secreted APPβ levels suggested that the elevated Aβ42 and Aβ40 levels were due to the attenuated Aβ degrading activity of BACE1. Notably, the amyloidolytic activity of BACE1 was impeded at lower BACE1 inhibitor concentrations compared to its amyloidogenic activity, thereby suggesting that the Aβ degrading activity of BACE1 was more sensitive to inhibition than its Aβ producing activity. Under endogenous BACE1 and APP levels, "low-dose" BACE1 inhibition affected both the Aβ producing and degrading activities of BACE1, i.e., significantly increased Aβ42/Aβ40 ratio and decreased Aβ34 levels, respectively. Further, we incubated recombinant BACE1 with synthetic Aβ peptides and found that BACE1 has a higher affinity for Aβ substrates over APP. In summary, our results suggest that stimulating BACE1's ADE activity and halting Aβ production without decreasing Aβ clearance could still be a promising therapeutic approach with new, yet to be developed, BACE1 modulators.
Keywords: Alzheimer’s disease; BACE1 amyloidolytic activity; amyloid clearance; amyloid production; secretase inhibitors.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures







References
-
- Masters C.L., Bateman R., Blennow K., Rowe C.C., Sperling R.A., Cummings J.L. Alzheimer's disease. Nat. Rev. Dis. Primers. 2015;1 - PubMed
-
- Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P., et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. - PubMed
-
- Jin S., Agerman K., Kolmodin K., Gustafsson E., Dahlqvist C., Jureus A., et al. Evidence for dimeric BACE-mediated APP processing. Biochem. Biophys. Res. Commun. 2010;393:21–27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

