Broadening the scope of multigene panel analysis for adult epilepsy patients
- PMID: 38946282
- PMCID: PMC11296137
- DOI: 10.1002/epi4.12993
Broadening the scope of multigene panel analysis for adult epilepsy patients
Abstract
Objective: Epilepsy is a suitable target for gene panel sequencing because a considerable portion of epilepsy is now explained by genetic components, especially in syndromic cases. However, previous gene panel studies on epilepsy have mostly focused on pediatric patients.
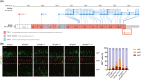
Methods: We enrolled adult epilepsy patients meeting any of the following criteria: family history of epilepsy, seizure onset age ≤ 19 years, neuronal migration disorder, and seizure freedom not achieved by dual anti-seizure medications. We sequenced the exonic regions of 211 epilepsy genes in these patients. To confirm the pathogenicity of a novel MTOR truncating variant, we electroporated vectors with different MTOR variants into developing mouse brains.
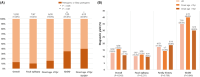
Results: A total of 92 probands and 4 affected relatives were tested, and the proportion of intellectual disability (ID) and/or developmental disability (DD) was 21.7%. As a result, twelve probands (13.0%) had pathogenic or likely pathogenic variants in the following genes or regions: DEPDC5, 15q12-q13 duplication (n = 2), SLC6A1, SYNGAP1, EEF1A2, LGI1, MTOR, KCNQ2, MEF2C, and TSC1 (n = 1). We confirmed the functional impact of a novel truncating mutation in the MTOR gene (c.7570C > T, p.Gln2524Ter) that disrupted neuronal migration in a mouse model. The diagnostic yield was higher in patients with ID/DD or childhood-onset seizures. We also identified additional candidate variants in 20 patients that could be reassessed by further studies.
Significance: Our findings underscore the clinical utility of gene panel sequencing in adult epilepsy patients suspected of having genetic etiology, especially those with ID/DD or early-onset seizures. Gene panel sequencing could not only lead to genetic diagnosis in a substantial portion of adult epilepsy patients but also inform more precise therapeutic decisions based on their genetic background.
Plain language summary: This study demonstrated the effectiveness of gene panel sequencing in adults with epilepsy, revealing pathogenic or likely pathogenic variants in 13.0% of patients. Higher diagnostic yields were observed in those with neurodevelopmental disorders or childhood-onset seizures. Additionally, we have shown that expanding genetic studies into adult patients would uncover new types of pathogenic variants for epilepsy, contributing to the advancement of precision medicine for individuals with epilepsy. In conclusion, our results highlight the practical value of employing gene panel sequencing in adult epilepsy patients, particularly when genetic etiology is clinically suspected.
Keywords: adult epilepsy; gene panel sequencing; next‐generation sequencing; precision medicine.
© 2024 The Author(s). Epilepsia Open published by Wiley Periodicals LLC on behalf of International League Against Epilepsy.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this study. We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Figures

References
-
- Borlot F, de Almeida BI, Combe SL, Andrade DM, Filloux FM, Myers KA. Clinical utility of multigene panel testing in adults with epilepsy and intellectual disability. Epilepsia. 2019;60:1661–1669. - PubMed
-
- Ellis CA, Petrovski S, Berkovic SF. Epilepsy genetics: clinical impacts and biological insights. Lancet Neurol. 2020;19:93–100. - PubMed
-
- Adams DR, Eng CM. Next‐generation sequencing to diagnose suspected genetic disorders. N Engl J Med. 2018;379:1353–1362. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

