Genomes and secondary metabolomes of Streptomyces spp. isolated from Leontopodium nivale ssp. alpinum
- PMID: 38946903
- PMCID: PMC11212599
- DOI: 10.3389/fmicb.2024.1408479
Genomes and secondary metabolomes of Streptomyces spp. isolated from Leontopodium nivale ssp. alpinum
Abstract
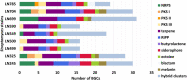
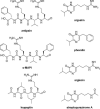
Bacterial endophytes dwelling in medicinal plants represent an as yet underexplored source of bioactive natural products with the potential to be developed into drugs against various human diseases. For the first time, several Streptomyces spp. were isolated from the rare and endangered traditional medicinal plant Leontopodium nivale ssp. alpinum, also known as Edelweiss. In the search for novel natural products, nine endophytic Streptomyces spp. from Edelweiss were investigated via genome sequencing and analysis, followed by fermentation in different media and investigation of secondary metabolomes. A total of 214 secondary metabolite biosynthetic gene clusters (BGCs), of which 35 are presumably unique, were identified by the bioinformatics tool antiSMASH in the genomes of these isolates. LC-MS analyses of the secondary metabolomes of these isolates revealed their potential to produce both known and presumably novel secondary metabolites, whereby most of the identified molecules could be linked to their cognate BGCs. This work sets the stage for further investigation of endophytic streptomycetes from Edelweiss aimed at the discovery and characterization of novel bioactive natural products.
Keywords: Edelweiss; Streptomyces; endophytes; genome mining; secondary metabolites.
Copyright © 2024 Malfent, Zehl, Kirkegaard, Oberhofer and Zotchev.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Secondary Metabolite Biosynthesis Potential of Streptomyces Spp. from the Rhizosphere of Leontopodium nivale Subsp. alpinum.ACS Omega. 2025 Feb 13;10(7):7163-7171. doi: 10.1021/acsomega.4c10476. eCollection 2025 Feb 25. ACS Omega. 2025. PMID: 40028056 Free PMC article.
-
Exploring Actinobacteria Associated With Rhizosphere and Endosphere of the Native Alpine Medicinal Plant Leontopodium nivale Subspecies alpinum.Front Microbiol. 2019 Nov 8;10:2531. doi: 10.3389/fmicb.2019.02531. eCollection 2019. Front Microbiol. 2019. PMID: 31781058 Free PMC article.
-
Biosynthetic Potential of the Endophytic Fungus Helotiales sp. BL73 Revealed via Compound Identification and Genome Mining.Appl Environ Microbiol. 2022 Mar 22;88(6):e0251021. doi: 10.1128/aem.02510-21. Epub 2022 Feb 2. Appl Environ Microbiol. 2022. PMID: 35108081 Free PMC article.
-
Endophytic Streptomyces: an underexplored source with potential for novel natural drug discovery and development.Arch Microbiol. 2024 Oct 22;206(11):442. doi: 10.1007/s00203-024-04169-z. Arch Microbiol. 2024. PMID: 39436470 Review.
-
Endophytic microbes from Nigerian ethnomedicinal plants: a potential source for bioactive secondary metabolites-a review.Bull Natl Res Cent. 2021;45(1):103. doi: 10.1186/s42269-021-00561-7. Epub 2021 Jun 8. Bull Natl Res Cent. 2021. PMID: 34121835 Free PMC article. Review.
Cited by
-
Secondary Metabolite Biosynthesis Potential of Streptomyces Spp. from the Rhizosphere of Leontopodium nivale Subsp. alpinum.ACS Omega. 2025 Feb 13;10(7):7163-7171. doi: 10.1021/acsomega.4c10476. eCollection 2025 Feb 25. ACS Omega. 2025. PMID: 40028056 Free PMC article.
-
The bacterial community of the freshwater bryozoan Cristatella Mucedo and its secondary metabolites production potential.Sci Rep. 2025 Aug 26;15(1):31456. doi: 10.1038/s41598-025-17084-0. Sci Rep. 2025. PMID: 40858791 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

