Targeting endothelial glycolytic reprogramming by tsRNA-1599 for ocular anti-angiogenesis therapy
- PMID: 38948065
- PMCID: PMC11209708
- DOI: 10.7150/thno.96946
Targeting endothelial glycolytic reprogramming by tsRNA-1599 for ocular anti-angiogenesis therapy
Abstract
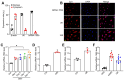
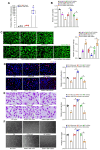
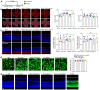
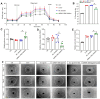
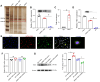
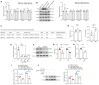
Rationale: Current treatments for ocular angiogenesis primarily focus on blocking the activity of vascular endothelial growth factor (VEGF), but unfavorable side effects and unsatisfactory efficacy remain issues. The identification of novel targets for anti-angiogenic treatment is still needed. Methods: We investigated the role of tsRNA-1599 in ocular angiogenesis using endothelial cells, a streptozotocin (STZ)-induced diabetic model, a laser-induced choroidal neovascularization model, and an oxygen-induced retinopathy model. CCK-8 assays, EdU assays, transwell assays, and matrigel assays were performed to assess the role of tsRNA-1599 in endothelial cells. Retinal digestion assays, Isolectin B4 (IB4) staining, and choroidal sprouting assays were conducted to evaluate the role of tsRNA-1599 in ocular angiogenesis. Transcriptomic analysis, metabolic analysis, RNA pull-down assays, and mass spectrometry were utilized to elucidate the mechanism underlying angiogenic effects mediated by tsRNA-1599. Results: tsRNA-1599 expression was up-regulated in experimental ocular angiogenesis models and endothelial cells in response to angiogenic stress. Silencing of tsRNA-1599 suppressed angiogenic effects in endothelial cells in vitro and inhibited pathological ocular angiogenesis in vivo. Mechanistically, tsRNA-1599 exhibited little effect on VEGF signaling but could cause reduced glycolysis and NAD+/NADH production in endothelial cells by regulating the expression of HK2 gene through interacting with YBX1, thus affecting endothelial effects. Conclusions: Targeting glycolytic reprogramming of endothelial cells by a tRNA-derived small RNA represents an exploitable therapeutic approach for ocular neovascular diseases.
Keywords: Angiogenesis; Endothelial metabolism; Glycolytic flux; Ocular neovascular disease; tsRNAs.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Eelen G, Treps L, Li X, Carmeliet P. Basic and therapeutic aspects of angiogenesis updated. Circ Res. 2020;127:310–29. - PubMed
-
- Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–57. - PubMed
-
- Li X, Carmeliet P. Targeting angiogenic metabolism in disease. Science. 2018;359:1335–6. - PubMed
-
- Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

