Dual activities of a silencing information regulator complex in yeast transcriptional regulation and DNA-damage response
- PMID: 38948145
- PMCID: PMC11211678
- DOI: 10.1002/mlf2.12108
Dual activities of a silencing information regulator complex in yeast transcriptional regulation and DNA-damage response
Abstract
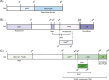
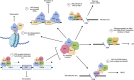
The Saccharomyces cerevisiae silencing information regulator (SIR) complex contains up to four proteins, namely Sir1, Sir2, Sir3, and Sir4. While Sir2 encodes a NAD-dependent histone deacetylase, other SIR proteins mainly function as structural and scaffold components through physical interaction with various proteins. The SIR complex displays different conformation and composition, including Sir2 homotrimer, Sir1-4 heterotetramer, Sir2-4 heterotrimer, and their derivatives, which recycle and relocate to different chromosomal regions. Major activities of the SIR complex are transcriptional silencing through chromosomal remodeling and modulation of DNA double-strand-break repair pathways. These activities allow the SIR complex to be involved in mating-type maintenance and switching, telomere and subtelomere gene silencing, promotion of nonhomologous end joining, and inhibition of homologous recombination, as well as control of cell aging. This review explores the potential link between epigenetic regulation and DNA damage response conferred by the SIR complex under various conditions aiming at understanding its roles in balancing cell survival and genomic stability in response to internal and environmental stresses. As core activities of the SIR complex are highly conserved in eukaryotes from yeast to humans, knowledge obtained in the yeast may apply to mammalian Sirtuin homologs and related diseases.
Keywords: DNA‐damage response; SIR complex; chromatin remodeling; transcriptional silencing; yeast.
© 2024 The Authors. mLife published by John Wiley & Sons Australia, Ltd on behalf of Institute of Microbiology, Chinese Academy of Sciences.
Figures


Similar articles
-
Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae.Nature. 1997 Aug 28;388(6645):900-3. doi: 10.1038/42288. Nature. 1997. PMID: 9278054
-
The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae.Genetics. 2016 Aug;203(4):1563-99. doi: 10.1534/genetics.112.145243. Genetics. 2016. PMID: 27516616 Free PMC article. Review.
-
Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation.Mol Cell Biol. 2002 Jun;22(12):4167-80. doi: 10.1128/MCB.22.12.4167-4180.2002. Mol Cell Biol. 2002. PMID: 12024030 Free PMC article.
-
Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths.Curr Biol. 1999 Jul 15;9(14):767-70. doi: 10.1016/s0960-9822(99)80339-x. Curr Biol. 1999. PMID: 10421582
-
The Sir2 family of protein deacetylases.Annu Rev Biochem. 2004;73:417-35. doi: 10.1146/annurev.biochem.73.011303.073651. Annu Rev Biochem. 2004. PMID: 15189148 Review.
References
-
- Oppikofer M, Kueng S, Gasser SM. SIR‐nucleosome interactions: structure‐function relationships in yeast silent chromatin. Gene. 2013;527:10–25. - PubMed
-
- Gross DS. Sir proteins as transcriptional silencers. Trends Biochem Sci. 2001;26:685–686. - PubMed
-
- Vaquero A, Scher M, Lee D, Erdjument‐Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
