PhysiCell Studio: a graphical tool to make agent-based modeling more accessible
- PMID: 38948511
- PMCID: PMC11211762
- DOI: 10.46471/gigabyte.128
PhysiCell Studio: a graphical tool to make agent-based modeling more accessible
Abstract
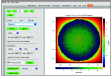
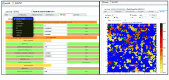
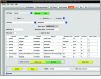
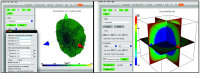
Defining a multicellular model can be challenging. There may be hundreds of parameters that specify the attributes and behaviors of objects. In the best case, the model will be defined using some format specification - a markup language - that will provide easy model sharing (and a minimal step toward reproducibility). PhysiCell is an open-source, physics-based multicellular simulation framework with an active and growing user community. It uses XML to define a model and, traditionally, users needed to manually edit the XML to modify the model. PhysiCell Studio is a tool to make this task easier. It provides a GUI that allows editing the XML model definition, including the creation and deletion of fundamental objects: cell types and substrates in the microenvironment. It also lets users build their model by defining initial conditions and biological rules, run simulations, and view results interactively. PhysiCell Studio has evolved over multiple workshops and academic courses in recent years, which has led to many improvements. There is both a desktop and cloud version. Its design and development has benefited from an active undergraduate and graduate research program. Like PhysiCell, the Studio is open-source software and contributions from the community are encouraged.
© The Author(s) 2024.
Conflict of interest statement
The authors declare they have no competing interests.
Figures









Update of
-
PhysiCell Studio: a graphical tool to make agent-based modeling more accessible.bioRxiv [Preprint]. 2023 Dec 14:2023.10.24.563727. doi: 10.1101/2023.10.24.563727. bioRxiv. 2023. Update in: GigaByte. 2024 Jun 19;2024:gigabyte128. doi: 10.46471/gigabyte.128. PMID: 37961612 Free PMC article. Updated. Preprint.
References
-
- Wilensky U. . NetLogo. 1999; http://ccl.northwestern.edu/netlogo. Accessed 1 September 2023.
-
- Pitt-Francis J, Pathmanathan P, Bernabeu MO et al. . Chaste: a test-driven approach to software development for biological modelling. Comput. Phys. Commun., 2009; 180: 2452–2471. doi:10.1016/j.cpc.2009.07.019. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
