This is a preprint.
Conditional frequency spectra as a tool for studying selection on complex traits in biobanks
- PMID: 38948697
- PMCID: PMC11212903
- DOI: 10.1101/2024.06.15.599126
Conditional frequency spectra as a tool for studying selection on complex traits in biobanks
Update in
-
Characterizing selection on complex traits through conditional frequency spectra.Genetics. 2025 Apr 17;229(4):iyae210. doi: 10.1093/genetics/iyae210. Genetics. 2025. PMID: 39691067
Abstract
Natural selection on complex traits is difficult to study in part due to the ascertainment inherent to genome-wide association studies (GWAS). The power to detect a trait-associated variant in GWAS is a function of frequency and effect size - but for traits under selection, the effect size of a variant determines the strength of selection against it, constraining its frequency. To account for GWAS ascertainment, we propose studying the joint distribution of allele frequencies across populations, conditional on the frequencies in the GWAS cohort. Before considering these conditional frequency spectra, we first characterized the impact of selection and non-equilibrium demography on allele frequency dynamics forwards and backwards in time. We then used these results to understand conditional frequency spectra under realistic human demography. Finally, we investigated empirical conditional frequency spectra for GWAS variants associated with 106 complex traits, finding compelling evidence for either stabilizing or purifying selection. Our results provide insight into polygenic score portability and other properties of variants ascertained with GWAS, highlighting the utility of conditional frequency spectra.
Figures


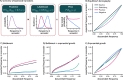


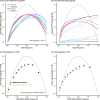
References
-
- Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A. A. E., Lee S. H., Robinson M. R., Perry J. R. B., Nolte I. M., van Vliet-Ostaptchouk J. V., et al. , 2015. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nature Genetics 47: 1114–1120. - PMC - PubMed
-
- Schoech A. P., Jordan D. M., Loh P.-R., Gazal S., O’Connor L. J., Balick D. J., Palamara P. F., Finucane H. K., Sunyaev S. R., and Price A. L., 2019. Quantification of frequency-dependent genetic architectures in 25 UK Biobank traits reveals action of negative selection. Nature Communications 10: 790. - PMC - PubMed
-
- Zeng J., de Vlaming R., Wu Y., Robinson M. R., Lloyd-Jones L. R., Yengo L., Yap C. X., Xue A., Sidorenko J., McRae A. F., et al. , 2018. Signatures of negative selection in the genetic architecture of human complex traits. Nature Genetics 50: 746–753. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
