This is a preprint.
Pathogenic Bacteroides fragilis strains can emerge from gut-resident commensals
- PMID: 38948766
- PMCID: PMC11213024
- DOI: 10.1101/2024.06.19.599758
Pathogenic Bacteroides fragilis strains can emerge from gut-resident commensals
Abstract
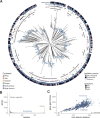
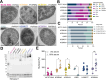
Bacteroides fragilis is a prominent member of the human gut microbiota, playing crucial roles in maintaining gut homeostasis and host health. Although it primarily functions as a beneficial commensal, B. fragilis can become pathogenic. To determine the genetic basis of its duality, we conducted a comparative genomic analysis of 813 B. fragilis strains, representing both commensal and pathogenic origins. Our findings reveal that pathogenic strains emerge across diverse phylogenetic lineages, due in part to rapid gene exchange and the adaptability of the accessory genome. We identified 16 phylogenetic groups, differentiated by genes associated with capsule composition, interspecies competition, and host interactions. A microbial genome-wide association study identified 44 genes linked to extra-intestinal survival and pathogenicity. These findings reveal how genomic diversity within commensal species can lead to the emergence of pathogenic traits, broadening our understanding of microbial evolution in the gut.
Conflict of interest statement
Conflict of Interest W.J.S’s current conflicts of interest are: Mirador Therapeutics (stock, employee, company officer, Ventyx Biosciences (stock, former employee), Prometheus Laboratory (board of directors), Shoreline Biosciences (stock, scientific advisory board), Forbion (consultant), Alimentiv (consultant). P.C.D. is an advisor and holds equity in Cybele, BileOmix, and Sirenas and a Scientific co-founder, and advisor and holds equity to Ometa, Enveda, and Arome with prior approval by UC-San Diego. P.C.D. also consulted for DSM animal health in 2023. R.K.’s current conflicts of interest are: Gencirq (stock and SAB member), DayTwo (consultant and SAB member), Cybele (stock and consultant), Biomesense (stock, consultant, SAB member), Micronoma (stock, SAB member, co-founder), and Biota (stock, co-founder).
Figures



References
-
- Yassour M., Vatanen T., Siljander H., Hämäläinen A.-M., Härkönen T., Ryhänen S.J., Franzosa E.A., Vlamakis H., Huttenhower C., Gevers D., et al. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatments on strain-level diversity and stability. Sci. Transl. Med. 8, 343ra81. 10.1126/scitranslmed.aad0917. - DOI - PMC - PubMed
-
- Kasper D.L., Onderdonk A.B., Polk B.F., Bartlett J.G. (1980). Surface Antigens as Virulence Factors in Infection with Bacteroides Fragilis. In Anaerobic Bacteria (Springer, Boston, MA: ). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
