This is a preprint.
Hierarchical global and local auxin signals coordinate cellular interdigitation in Arabidopsis
- PMID: 38948792
- PMCID: PMC11212924
- DOI: 10.1101/2024.06.17.599171
Hierarchical global and local auxin signals coordinate cellular interdigitation in Arabidopsis
Abstract
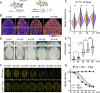
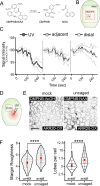
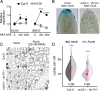
The development of multicellular tissues requires both local and global coordination of cell polarization, however, the mechanisms underlying their interplay are poorly understood. In Arabidopsis, leaf epidermal pavement cells (PC) develop a puzzle-piece shape locally coordinated through apoplastic auxin signaling. Here we show auxin also globally coordinates interdigitation by activating the TIR1/AFB-dependent nuclear signaling pathway. This pathway promotes a transient maximum of auxin at the cotyledon tip, which then moves across the leaf activating local PC polarization, as demonstrated by locally uncaged auxin globally rescuing defects in tir1;afb1;afb2;afb4;afb5 mutant but not in tmk1;tmk2;tmk3;tmk4 mutants. Our findings show that hierarchically integrated global and local auxin signaling systems, which respectively depend on TIR1/AFB-dependent gene transcription in the nucleus and TMK-mediated rapid activation of ROP GTPases at the cell surface, control PC interdigitation patterns in Arabidopsis cotyledons, revealing a mechanism for coordinating a local cellular process with the development of whole tissues.
Keywords: Cell polarity; TIR1/AFBs; TMK; auxin transport; global coordination; local coordination; pavement cell morphogenesis.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
