Atomistic insights into the reentrant phase-transitions in polyuracil and polylysine mixtures
- PMID: 38949285
- PMCID: PMC11378353
- DOI: 10.1063/5.0206190
Atomistic insights into the reentrant phase-transitions in polyuracil and polylysine mixtures
Abstract
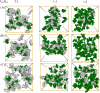
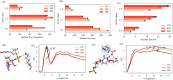
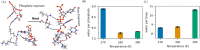
The phase separation of protein and RNA mixtures underpins the assembly and regulation of numerous membraneless organelles in cells. The ubiquity of protein-RNA condensates in cellular regulatory processes is in part due to their sensitivity to RNA concentration, which affects their physical properties and stability. Recent experiments with poly-cationic peptide-RNA mixtures have revealed closed-loop phase diagrams featuring lower and upper critical solution temperatures. These diagrams indicate reentrant phase transitions shaped by biomolecular interactions and entropic forces such as solvent and ion reorganization. We employed atomistic simulations to study mixtures with various RNA-polylysine stoichiometries and temperatures to elucidate the microscopic driving forces behind reentrant phase transitions in protein-RNA mixtures. Our findings reveal an intricate interplay between hydration, ion condensation, and specific RNA-polylysine hydrogen bonding, resulting in distinct stoichiometry-dependent phase equilibria governing stabilities and structures of the condensate phase. Our simulations show that reentrant transitions are accompanied by desolvation around the phosphate groups of RNA, with increased contacts between phosphate and lysine side chains. In RNA-rich systems at lower temperatures, RNA molecules can form an extensive pi-stacking and hydrogen bond network, leading to percolation. In protein-rich systems, no such percolation-induced transitions are observed. Furthermore, we assessed the performance of three prominent water force fields-Optimal Point Charge (OPC), TIP4P-2005, and TIP4P-D-in capturing reentrant phase transitions. OPC provided a superior balance of interactions, enabling effective capture of reentrant transitions and accurate characterization of changes in solvent reorganization. This study offers atomistic insights into the nature of reentrant phase transitions using simple model peptide and nucleotide mixtures. We believe that our results are broadly applicable to larger classes of peptide-RNA mixtures exhibiting reentrant phase transitions.
© 2024 Author(s). Published under an exclusive license by AIP Publishing.
Conflict of interest statement
The authors have no conflicts to disclose.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

