NAPRT Silencing in FH-Deficient Renal Cell Carcinoma Confers Therapeutic Vulnerabilities via NAD+ Depletion
- PMID: 38949523
- PMCID: PMC11445649
- DOI: 10.1158/1541-7786.MCR-23-1003
NAPRT Silencing in FH-Deficient Renal Cell Carcinoma Confers Therapeutic Vulnerabilities via NAD+ Depletion
Abstract
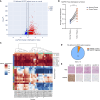
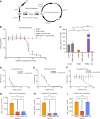
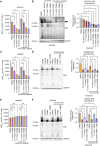
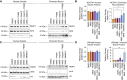
Hereditary leiomyomatosis and renal cell carcinoma (HLRCC) is caused by loss of function mutations in fumarate hydratase (FH) and results in an aggressive subtype of renal cell carcinoma with limited treatment options. Loss of FH leads to accumulation of fumarate, an oncometabolite that disrupts multiple cellular processes and drives tumor progression. High levels of fumarate inhibit alpha ketoglutarate-dependent dioxygenases, including the ten-eleven translocation (TET) enzymes, and can lead to global DNA hypermethylation. Here, we report patterns of hypermethylation in FH-mutant cell lines and tumor samples are associated with the silencing of nicotinate phosphoribosyl transferase (NAPRT), a rate-limiting enzyme in the Preiss-Handler pathway of NAD+ biosynthesis, in a subset of HLRCC cases. NAPRT is hypermethylated at a CpG island in the promoter in cell line models and patient samples, resulting in loss of NAPRT expression. We find that FH-deficient RCC models with loss of NAPRT expression, as well as other oncometabolite-producing cancer models that silence NAPRT, are extremely sensitive to nicotinamide phosphoribosyl transferase inhibitors (NAMPTi). NAPRT silencing was also associated with synergistic tumor cell killing with PARP inhibitors and NAMPTis, which was associated with effects on PAR-mediated DNA repair. Overall, our findings indicate that NAPRT silencing can be targeted in oncometabolite-producing cancers and elucidates how oncometabolite-associated hypermethylation can impact diverse cellular processes and lead to therapeutically relevant vulnerabilities in cancer cells. Implications: NAPRT is a novel biomarker for targeting NAD+ metabolism in FH-deficient HLRCCs with NAMPTis alone and targeting DNA repair processes with the combination of NAMPTis and PARP inhibitors.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
K.J. Noronha reports grants from NIH (R01CA21543-05, R.S. Bindra; NRSA F31, K.J. Noronha) and Department of Defense (DOD; W81XWH-22-1-0549, B. Shuch and R.S. Bindra) during the conduct of the study and personal fees from AtlasXomics, Inc., outside the submitted work. K.N. Lucas reports grants from NIH (R01CA21543-05, R.S. Bindra) and DOD (W81XWH-22-1-0549, B. Shuch and R.S. Bindra) during the conduct of the study and other support from York Analytical Laboratories outside the submitted work. S. Friedman reports grants from NIH (R01CA21543-05, R.S. Bindra) and DOD (W81XWH-22-1-0549, B. Shuch and R.S. Bindra) during the conduct of the study. M.A. Murray reports grants from NIH NRSA F31 outside the submitted work. S. Liu reports grants from Yale College First-Year Research Fellowship during the conduct of the study. J. Spurrier reports employment with Alphina Therapeutics. M. Raponi reports a patent for Anti-NAPRT Antibodies and Methods of Use pending to Alphina Therapeutics. J.C. Vasquez reports grants from NIH K08, Robert Wood Johnson Harold Amos Medical Faculty Development Program, Doris Duke Charitable Foundation, and American Cancer Society Institutional Research Grant during the conduct of the study. No disclosures were reported by the other authors.
Figures


References
-
- Srinivasan R, Gurram S, Harthy MA, Singer EA, Sidana A, Shuch BM, et al. . Results from a phase II study of bevacizumab and erlotinib in subjects with advanced hereditary leiomyomatosis and renal cell cancer (HLRCC) or sporadic papillary renal cell cancer. J Clin Oncol 2020;38(15_Suppl):5004.
-
- Carril-Ajuria L, Colomba E, Cerbone L, Romero-Ferreiro C, Crouzet L, Laguerre B, et al. . Response to systemic therapy in fumarate hydratase-deficient renal cell carcinoma. Eur J Cancer 2021;151:106–14. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- F31 CA261129/CA/NCI NIH HHS/United States
- K08 CA258796/CA/NCI NIH HHS/United States
- 1-K08 CA258796-01/National Cancer Institute (NCI)
- Robert Wood Johnson Foundation (RWJF)
- UL1 TR001863/TR/NCATS NIH HHS/United States
- R01 CA215453/CA/NCI NIH HHS/United States
- IRG-21-132-60-IRG/American Cancer Society (ACS)
- F31CA260794/National Cancer Institute (NCI)
- W81XWH-22-1-0549/U.S. Department of Defense (DOD)
- 2015216/Doris Duke Charitable Foundation (DDCF)
- F31 CA260794/CA/NCI NIH HHS/United States
- R01CA21543-05/National Cancer Institute (NCI)
- F31CA261129/National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

