A fluorogenic complementation tool kit for interrogating lipid droplet-organelle interaction
- PMID: 38949658
- PMCID: PMC11215687
- DOI: 10.1083/jcb.202311126
A fluorogenic complementation tool kit for interrogating lipid droplet-organelle interaction
Abstract
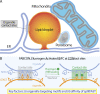
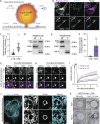
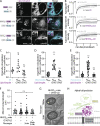
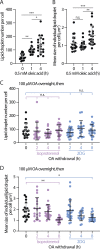
Contact sites between lipid droplets and other organelles are essential for cellular lipid and energy homeostasis upon metabolic demands. Detection of these contact sites at the nanometer scale over time in living cells is challenging. We developed a tool kit for detecting contact sites based on fluorogen-activated bimolecular complementation at CONtact sites, FABCON, using a reversible, low-affinity split fluorescent protein, splitFAST. FABCON labels contact sites with minimal perturbation to organelle interaction. Via FABCON, we quantitatively demonstrated that endoplasmic reticulum (ER)- and mitochondria (mito)-lipid droplet contact sites are dynamic foci in distinct metabolic conditions, such as during lipid droplet biogenesis and consumption. An automated analysis pipeline further classified individual contact sites into distinct subgroups based on size, likely reflecting differential regulation and function. Moreover, FABCON is generalizable to visualize a repertoire of organelle contact sites including ER-mito. Altogether, FABCON reveals insights into the dynamic regulation of lipid droplet-organelle contact sites and generates new hypotheses for further mechanistical interrogation during metabolic regulation.
© 2024 Li et al.
Conflict of interest statement
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. A. Tebo reported a patent to US20220169682A1 pending. No other disclosures were reported.
Figures





Update of
-
A fluorogenic complementation tool kit for interrogating lipid droplet-organelle interaction.bioRxiv [Preprint]. 2023 Nov 29:2023.11.29.569289. doi: 10.1101/2023.11.29.569289. bioRxiv. 2023. Update in: J Cell Biol. 2024 Sep 2;223(9):e202311126. doi: 10.1083/jcb.202311126. PMID: 38076863 Free PMC article. Updated. Preprint.
Similar articles
-
A fluorogenic complementation tool kit for interrogating lipid droplet-organelle interaction.bioRxiv [Preprint]. 2023 Nov 29:2023.11.29.569289. doi: 10.1101/2023.11.29.569289. bioRxiv. 2023. Update in: J Cell Biol. 2024 Sep 2;223(9):e202311126. doi: 10.1083/jcb.202311126. PMID: 38076863 Free PMC article. Updated. Preprint.
-
Long-term live-cell lipid droplet-targeted biosensor development for nanoscopic tracking of lipid droplet-mitochondria contact sites.Theranostics. 2021 Jun 22;11(16):7767-7778. doi: 10.7150/thno.59848. eCollection 2021. Theranostics. 2021. PMID: 34335963 Free PMC article.
-
Spatial compartmentalization of lipid droplet biogenesis.Biochim Biophys Acta Mol Cell Biol Lipids. 2020 Jan;1865(1):158499. doi: 10.1016/j.bbalip.2019.07.008. Epub 2019 Jul 25. Biochim Biophys Acta Mol Cell Biol Lipids. 2020. PMID: 31352131 Free PMC article. Review.
-
Perspectives on Mitochondria-ER and Mitochondria-Lipid Droplet Contact in Hepatocytes and Hepatic Lipid Metabolism.Cells. 2021 Sep 1;10(9):2273. doi: 10.3390/cells10092273. Cells. 2021. PMID: 34571924 Free PMC article. Review.
-
Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility.Elife. 2019 Feb 11;8:e43561. doi: 10.7554/eLife.43561. Elife. 2019. PMID: 30741634 Free PMC article.
Cited by
-
Nondisruptive inducible labeling of ER-membrane contact sites using the Lamin B receptor.PLoS Biol. 2025 Jul 10;23(7):e3003249. doi: 10.1371/journal.pbio.3003249. eCollection 2025 Jul. PLoS Biol. 2025. PMID: 40638559 Free PMC article.
-
A tunable and versatile chemogenetic near-infrared fluorescent reporter.Nat Commun. 2025 Mar 16;16(1):2594. doi: 10.1038/s41467-025-58017-9. Nat Commun. 2025. PMID: 40091099 Free PMC article.
-
Key challenges and recommendations for defining organelle membrane contact sites.Nat Rev Mol Cell Biol. 2025 Jun 23. doi: 10.1038/s41580-025-00864-x. Online ahead of print. Nat Rev Mol Cell Biol. 2025. PMID: 40550870 Review.
-
Imaging and proteomics toolkits for studying organelle contact sites.Front Cell Dev Biol. 2024 Sep 24;12:1466915. doi: 10.3389/fcell.2024.1466915. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39381373 Free PMC article. Review.
-
Imaging interorganelle contacts at a glance.J Cell Sci. 2024 Oct 15;137(20):jcs262020. doi: 10.1242/jcs.262020. Epub 2024 Oct 23. J Cell Sci. 2024. PMID: 39440475 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

