IL-10 suppresses T cell expansion while promoting tissue-resident memory cell formation during SARS-CoV-2 infection in rhesus macaques
- PMID: 38950078
- PMCID: PMC11244803
- DOI: 10.1371/journal.ppat.1012339
IL-10 suppresses T cell expansion while promoting tissue-resident memory cell formation during SARS-CoV-2 infection in rhesus macaques
Abstract
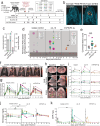
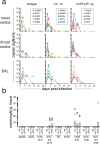
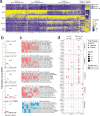
The regulation of inflammatory responses and pulmonary disease during SARS-CoV-2 infection is incompletely understood. Here we examine the roles of the prototypic pro- and anti-inflammatory cytokines IFNγ and IL-10 using the rhesus macaque model of mild COVID-19. We find that IFNγ drives the development of 18fluorodeoxyglucose (FDG)-avid lesions in the lungs as measured by PET/CT imaging but is not required for suppression of viral replication. In contrast, IL-10 limits the duration of acute pulmonary lesions, serum markers of inflammation and the magnitude of virus-specific T cell expansion but does not impair viral clearance. We also show that IL-10 induces the subsequent differentiation of virus-specific effector T cells into CD69+CD103+ tissue resident memory cells (Trm) in the airways and maintains Trm cells in nasal mucosal surfaces, highlighting an unexpected role for IL-10 in promoting airway memory T cells during SARS-CoV-2 infection of macaques.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
A.S. is a consultant for Gritstone Bio, Flow Pharma, Moderna, AstraZeneca, Qiagen, Avalia, Fortress, Gilead, Sanofi, Merck, RiverVest, MedaCorp, Turnstone, NA Vaccine Institute, Gerson Lehrman Group and Guggenheim. LJI has filed for patent protection for various aspects of T cell epitope and vaccine design work. All other authors have no competing interests to disclose.
Figures



Update of
-
IL-10 suppresses T cell expansion while promoting tissue-resident memory cell formation during SARS-CoV-2 infection in rhesus macaques.bioRxiv [Preprint]. 2022 Sep 15:2022.09.13.507852. doi: 10.1101/2022.09.13.507852. bioRxiv. 2022. Update in: PLoS Pathog. 2024 Jul 1;20(7):e1012339. doi: 10.1371/journal.ppat.1012339. PMID: 36172119 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

