HIV-1 Vpr combats the PU.1-driven antiviral response in primary human macrophages
- PMID: 38951492
- PMCID: PMC11217462
- DOI: 10.1038/s41467-024-49635-w
HIV-1 Vpr combats the PU.1-driven antiviral response in primary human macrophages
Abstract
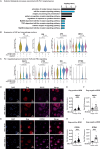
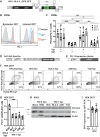
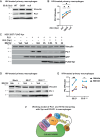
HIV-1 Vpr promotes efficient spread of HIV-1 from macrophages to T cells by transcriptionally downmodulating restriction factors that target HIV-1 Envelope protein (Env). Here we find that Vpr induces broad transcriptomic changes by targeting PU.1, a transcription factor necessary for expression of host innate immune response genes, including those that target Env. Consistent with this, we find silencing PU.1 in infected macrophages lacking Vpr rescues Env. Vpr downmodulates PU.1 through a proteasomal degradation pathway that depends on physical interactions with PU.1 and DCAF1, a component of the Cul4A E3 ubiquitin ligase. The capacity for Vpr to target PU.1 is highly conserved across primate lentiviruses. In addition to impacting infected cells, we find that Vpr suppresses expression of innate immune response genes in uninfected bystander cells, and that virion-associated Vpr can degrade PU.1. Together, we demonstrate Vpr counteracts PU.1 in macrophages to blunt antiviral immune responses and promote viral spread.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
HIV-1 Vpr combats the PU.1-driven antiviral response in primary human macrophages.bioRxiv [Preprint]. 2024 Feb 27:2023.03.21.533528. doi: 10.1101/2023.03.21.533528. bioRxiv. 2024. Update in: Nat Commun. 2024 Jun 29;15(1):5514. doi: 10.1038/s41467-024-49635-w. PMID: 36993393 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- T32 GM008353/GM/NIGMS NIH HHS/United States
- F31AI15504/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
- R01AI149669/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
- R21 AI132379/AI/NIAID NIH HHS/United States
- R61 DA059916/DA/NIDA NIH HHS/United States
- R21AI32379/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
- 1R61DA059916-01/U.S. Department of Health & Human Services | NIH | National Institute on Drug Abuse (NIDA)
- F31AI125090-01/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
- R25 GM086262/GM/NIGMS NIH HHS/United States
- R01 AI149669/AI/NIAID NIH HHS/United States
- T32GM007315/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- F31 AI125090/AI/NIAID NIH HHS/United States
- 5T32GM008353-27/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- T32 GM007315/GM/NIGMS NIH HHS/United States

