The chromatin regulator Ankrd11 controls cardiac neural crest cell-mediated outflow tract remodeling and heart function
- PMID: 38951500
- PMCID: PMC11217281
- DOI: 10.1038/s41467-024-48955-1
The chromatin regulator Ankrd11 controls cardiac neural crest cell-mediated outflow tract remodeling and heart function
Abstract
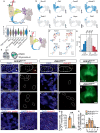
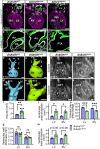
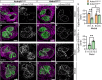
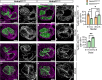
ANKRD11 (Ankyrin Repeat Domain 11) is a chromatin regulator and a causative gene for KBG syndrome, a rare developmental disorder characterized by multiple organ abnormalities, including cardiac defects. However, the role of ANKRD11 in heart development is unknown. The neural crest plays a leading role in embryonic heart development, and its dysfunction is implicated in congenital heart defects. We demonstrate that conditional knockout of Ankrd11 in the murine embryonic neural crest results in persistent truncus arteriosus, ventricular dilation, and impaired ventricular contractility. We further show these defects occur due to aberrant cardiac neural crest cell organization leading to outflow tract septation failure. Lastly, knockout of Ankrd11 in the neural crest leads to impaired expression of various transcription factors, chromatin remodelers and signaling pathways, including mTOR, BMP and TGF-β in the cardiac neural crest cells. In this work, we identify Ankrd11 as a regulator of neural crest-mediated heart development and function.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: J.M. is an employee of Vizgen. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- P34136/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- 3925/Women and Children's Health Research Institute (Women & Children's Health Research Institute)
- 20240101/Barncancerfonden (Swedish Childhood Cancer Foundation)
- P 34136/FWF_/Austrian Science Fund FWF/Austria
- 2023-02161/Vetenskapsrådet (Swedish Research Council)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

