PEX3 promotes regenerative repair after myocardial injury in mice through facilitating plasma membrane localization of ITGB3
- PMID: 38951640
- PMCID: PMC11217276
- DOI: 10.1038/s42003-024-06483-0
PEX3 promotes regenerative repair after myocardial injury in mice through facilitating plasma membrane localization of ITGB3
Abstract
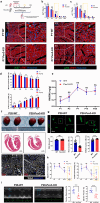
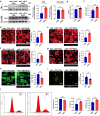
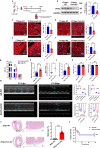
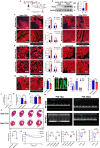
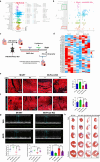
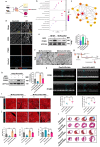
The peroxisome is a versatile organelle that performs diverse metabolic functions. PEX3, a critical regulator of the peroxisome, participates in various biological processes associated with the peroxisome. Whether PEX3 is involved in peroxisome-related redox homeostasis and myocardial regenerative repair remains elusive. We investigate that cardiomyocyte-specific PEX3 knockout (Pex3-KO) results in an imbalance of redox homeostasis and disrupts the endogenous proliferation/development at different times and spatial locations. Using Pex3-KO mice and myocardium-targeted intervention approaches, the effects of PEX3 on myocardial regenerative repair during both physiological and pathological stages are explored. Mechanistically, lipid metabolomics reveals that PEX3 promotes myocardial regenerative repair by affecting plasmalogen metabolism. Further, we find that PEX3-regulated plasmalogen activates the AKT/GSK3β signaling pathway via the plasma membrane localization of ITGB3. Our study indicates that PEX3 may represent a novel therapeutic target for myocardial regenerative repair following injury.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

