Mitochondrial genetics through the lens of single-cell multi-omics
- PMID: 38951641
- PMCID: PMC11260401
- DOI: 10.1038/s41588-024-01794-8
Mitochondrial genetics through the lens of single-cell multi-omics
Abstract
Mitochondria carry their own genetic information encoding for a subset of protein-coding genes and translational machinery essential for cellular respiration and metabolism. Despite its small size, the mitochondrial genome, its natural genetic variation and molecular phenotypes have been challenging to study using bulk sequencing approaches, due to its variation in cellular copy number, non-Mendelian modes of inheritance and propensity for mutations. Here we highlight emerging strategies designed to capture mitochondrial genetic variation across individual cells for lineage tracing and studying mitochondrial genetics in primary human cells and clinical specimens. We review recent advances surrounding single-cell mitochondrial genome sequencing and its integration with functional genomic readouts, including leveraging somatic mitochondrial DNA mutations as clonal markers that can resolve cellular population dynamics in complex human tissues. Finally, we discuss how single-cell whole mitochondrial genome sequencing approaches can be utilized to investigate mitochondrial genetics and its contribution to cellular heterogeneity and disease.
© 2024. Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The Broad Institute has filed for patents relating to the use of technologies described in this paper where C.A.L. and L.S.L., are named inventors (US patent applications 17/251,451 and 17/928,696). C.A.L. and L.S.L. are consultants to Cartography Biosciences. L.N. declares no competing interests.
Figures



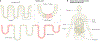
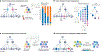
References
-
- Wallace DC Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76, 781–821 (2007). - PubMed
-
- Stewart JB & Chinnery PF The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 16, 530–542 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- n/a/Hector Fellow Academy
- STA 1586/5-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB1588/Deutsche Forschungsgemeinschaft (German Research Foundation)
- P30 CA008748/CA/NCI NIH HHS/United States
- K99 HG012579/HG/NHGRI NIH HHS/United States
- LU 2336/3-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Young Investigator/European Molecular Biology Organization (EMBO)
- UM1 HG012076/HG/NHGRI NIH HHS/United States
- LU 2336/2-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- R00 HG012579/HG/NHGRI NIH HHS/United States
- LU 2336/6-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- LU 2336/9-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- HG012579/U.S. Department of Health & Human Services | NIH | National Human Genome Research Institute (NHGRI)
LinkOut - more resources
Full Text Sources
Medical

