Understanding the genetic complexity of puberty timing across the allele frequency spectrum
- PMID: 38951643
- PMCID: PMC11250262
- DOI: 10.1038/s41588-024-01798-4
Understanding the genetic complexity of puberty timing across the allele frequency spectrum
Erratum in
-
Publisher Correction: Understanding the genetic complexity of puberty timing across the allele frequency spectrum.Nat Genet. 2024 Aug;56(8):1763-1764. doi: 10.1038/s41588-024-01857-w. Nat Genet. 2024. PMID: 38982295 Free PMC article. No abstract available.
Abstract
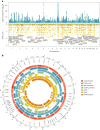
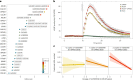
Pubertal timing varies considerably and is associated with later health outcomes. We performed multi-ancestry genetic analyses on ~800,000 women, identifying 1,080 signals for age at menarche. Collectively, these explained 11% of trait variance in an independent sample. Women at the top and bottom 1% of polygenic risk exhibited ~11 and ~14-fold higher risks of delayed and precocious puberty, respectively. We identified several genes harboring rare loss-of-function variants in ~200,000 women, including variants in ZNF483, which abolished the impact of polygenic risk. Variant-to-gene mapping approaches and mouse gonadotropin-releasing hormone neuron RNA sequencing implicated 665 genes, including an uncharacterized G-protein-coupled receptor, GPR83, which amplified the signaling of MC3R, a key nutritional sensor. Shared signals with menopause timing at genes involved in DNA damage response suggest that the ovarian reserve might signal centrally to trigger puberty. We also highlight body size-dependent and independent mechanisms that potentially link reproductive timing to later life disease.
© 2024. The Author(s).
Conflict of interest statement
J.R.B.P. and E.J.G. are employees/shareholders of Insmed. J.R.B.P. also receives research funding from GSK and consultancy fees from WW International. Y. Zhao is a UK University worker at GSK. D.L.C. is an employee and shareholder of GSK. J.P.B. is employed by GSK. I.S.F. has consulted for a number of companies developing weight loss drugs including Eli Lilly, Novo Nordisk and Rhythm Pharmaceuticals. D.J.T. is employed by Genomics PLC. E.T. is employed by Pfizer. D.A.L. has received support from Roche Diagnostics and Medtronic for work unrelated to the research in this paper. T.D.S. is cofounder and stakeholder of Zoe Global. P.A.F. conducts research funded by Amgen, Novartis and Pfizer, and he received Honoraria from Roche, Novartis and Pfizer. M.W.B. conducts research funded by Amgen, Novartis and Pfizer. M.I.M. is currently an employee of Genentech and a holder of Roche stock. The other authors declare no competing interests.
Figures




Update of
-
Understanding the genetic complexity of puberty timing across the allele frequency spectrum.medRxiv [Preprint]. 2023 Jun 20:2023.06.14.23291322. doi: 10.1101/2023.06.14.23291322. medRxiv. 2023. Update in: Nat Genet. 2024 Jul;56(7):1397-1411. doi: 10.1038/s41588-024-01798-4. PMID: 37503126 Free PMC article. Updated. Preprint.

