TRIM65 deficiency alleviates renal fibrosis through NUDT21-mediated alternative polyadenylation
- PMID: 38951701
- PMCID: PMC11519343
- DOI: 10.1038/s41418-024-01336-z
TRIM65 deficiency alleviates renal fibrosis through NUDT21-mediated alternative polyadenylation
Abstract
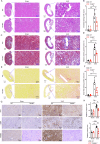
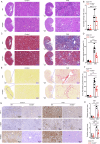
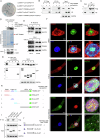
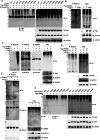
Chronic kidney disease (CKD) is a major global health concern and the third leading cause of premature death. Renal fibrosis is the primary process driving the progression of CKD, but the mechanisms behind it are not fully understood, making treatment options limited. Here, we find that the E3 ligase TRIM65 is a positive regulator of renal fibrosis. Deletion of TRIM65 results in a reduction of pathological lesions and renal fibrosis in mouse models of kidney fibrosis induced by unilateral ureteral obstruction (UUO)- and folic acid. Through screening with a yeast-hybrid system, we identify a new interactor of TRIM65, the mammalian cleavage factor I subunit CFIm25 (NUDT21), which plays a crucial role in fibrosis through alternative polyadenylation (APA). TRIM65 interacts with NUDT21 to induce K48-linked polyubiquitination of lysine 56 and proteasomal degradation, leading to the inhibition of TGF-β1-mediated SMAD and ERK1/2 signaling pathways. The degradation of NUDT21 subsequently altered the length and sequence content of the 3'UTR (3'UTR-APA) of several pro-fibrotic genes including Col1a1, Fn-1, Tgfbr1, Wnt5a, and Fzd2. Furthermore, reducing NUDT21 expression via hydrodynamic renal pelvis injection of adeno-associated virus 9 (AAV9) exacerbated UUO-induced renal fibrosis in the normal mouse kidneys and blocked the protective effect of TRIM65 deletion. These findings suggest that TRIM65 promotes renal fibrosis by regulating NUDT21-mediated APA and highlight TRIM65 as a potential target for reducing renal fibrosis in CKD patients.
© 2024. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Romagnani P, Remuzzi G, Glassock R, Levin A, Jager KJ, Tonelli M, et al. Chronic kidney disease. Nat Rev Dis Prim. 2017;3:17088. - PubMed
-
- Carney EF. The impact of chronic kidney disease on global health. Nat Rev Nephrol. 2020;16:251. - PubMed
-
- Glassock RJ, Warnock DG, Delanaye P. The global burden of chronic kidney disease: estimates, variability and pitfalls. Nat Rev Nephrol. 2017;13:104–14. - PubMed
MeSH terms
Substances
Grants and funding
- 20212ACB216005/Natural Science Foundation of Jiangxi Province (Jiangxi Province Natural Science Foundation)
- 20224BAB206007/Natural Science Foundation of Jiangxi Province (Jiangxi Province Natural Science Foundation)
- 20224ACB216013/Natural Science Foundation of Jiangxi Province (Jiangxi Province Natural Science Foundation)
- 20212BAB206086/Natural Science Foundation of Jiangxi Province (Jiangxi Province Natural Science Foundation)
- 82160133/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Miscellaneous

