DHPS-Mediated Hypusination Regulates METTL3 Self-m6A-Methylation Modification to Promote Melanoma Proliferation and the Development of Novel Inhibitors
- PMID: 38952061
- PMCID: PMC11434010
- DOI: 10.1002/advs.202402450
DHPS-Mediated Hypusination Regulates METTL3 Self-m6A-Methylation Modification to Promote Melanoma Proliferation and the Development of Novel Inhibitors
Abstract
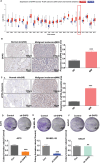
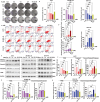
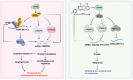
Discovering new treatments for melanoma will benefit human health. The mechanism by which deoxyhypusine synthase (DHPS) promotes melanoma development remains elucidated. Multi-omics studies have revealed that DHPS regulates m6A modification and maintains mRNA stability in melanoma cells. Mechanistically, DHPS activates the hypusination of eukaryotic translation initiation factor 5A (eIF5A) to assist METTL3 localizing on its mRNA for m6A modification, then promoting METTL3 expression. Structure-based design, synthesis, and activity screening yielded the hit compound GL-1 as a DHPS inhibitor. Notably, GL-1 directly inhibits DHPS binding to eIF5A, whereas GC-7 cannot. Based on the clarification of the mode of action of GL-1 on DHPS, it is found that GL-1 can promote the accumulation of intracellular Cu2+ to induce apoptosis, and antibody microarray analysis shows that GL-1 inhibits the expression of several cytokines. GL-1 shows promising antitumor activity with good bioavailability in a xenograft tumor model. These findings clarify the molecular mechanisms by which DHPS regulates melanoma proliferation and demonstrate the potential of GL-1 for clinical melanoma therapy.
Keywords: DHPS; eIF5A‐Hypusine; m6A; melanoma.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
