Identification of the proteolytic signature in CVB3-infected cells
- PMID: 38953667
- PMCID: PMC11265341
- DOI: 10.1128/jvi.00498-24
Identification of the proteolytic signature in CVB3-infected cells
Abstract
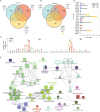
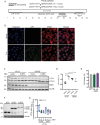
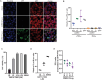
Coxsackievirus B3 (CVB3) encodes proteinases that are essential for processing of the translated viral polyprotein. Viral proteinases also target host proteins to manipulate cellular processes and evade innate antiviral responses to promote replication and infection. While some host protein substrates of the CVB3 3C and 2A cysteine proteinases have been identified, the full repertoire of targets is not known. Here, we utilize an unbiased quantitative proteomics-based approach termed terminal amine isotopic labeling of substrates (TAILS) to conduct a global analysis of CVB3 protease-generated N-terminal peptides in both human HeLa and mouse cardiomyocyte (HL-1) cell lines infected with CVB3. We identified >800 proteins that are cleaved in CVB3-infected HeLa and HL-1 cells including the viral polyprotein, known substrates of viral 3C proteinase such as PABP, DDX58, and HNRNPs M, K, and D and novel cellular proteins. Network and GO-term analysis showed an enrichment in biological processes including immune response and activation, RNA processing, and lipid metabolism. We validated a subset of candidate substrates that are cleaved under CVB3 infection and some are direct targets of 3C proteinase in vitro. Moreover, depletion of a subset of TAILS-identified target proteins decreased viral yield. Characterization of two target proteins showed that expression of 3Cpro-targeted cleaved fragments of emerin and aminoacyl-tRNA synthetase complex-interacting multifunctional protein 2 modulated autophagy and the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway, respectively. The comprehensive identification of host proteins targeted during virus infection provides insights into the cellular pathways manipulated to facilitate infection.
Importance: RNA viruses encode proteases that are responsible for processing viral proteins into their mature form. Viral proteases also target and cleave host cellular proteins; however, the full catalog of these target proteins is incomplete. We use a technique called terminal amine isotopic labeling of substrates (TAILS), an N-terminomics to identify host proteins that are cleaved under virus infection. We identify hundreds of cellular proteins that are cleaved under infection, some of which are targeted directly by viral protease. Revealing these target proteins provides insights into the host cellular pathways and antiviral signaling factors that are modulated to promote virus infection and potentially leading to virus-induced pathogenesis.
Keywords: AIMP2; EMD; N-terminomics; degradomics; picornavirus; protease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Collaborators GBoDS . 2015. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the global burden of disease study 2013. Lancet 386:743–800. doi:10.1016/S0140-6736(15)60692-4 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

