Single-cell RNA-Seq analysis reveals cell subsets and gene signatures associated with rheumatoid arthritis disease activity
- PMID: 38954480
- PMCID: PMC11343607
- DOI: 10.1172/jci.insight.178499
Single-cell RNA-Seq analysis reveals cell subsets and gene signatures associated with rheumatoid arthritis disease activity
Abstract
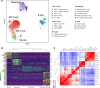
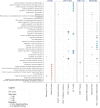
Rheumatoid arthritis (RA) management leans toward achieving remission or low disease activity. In this study, we conducted single-cell RNA sequencing (scRNA-Seq) of peripheral blood mononuclear cells (PBMCs) from 36 individuals (18 patients with RA and 18 matched controls, accounting for age, sex, race, and ethnicity), to identify disease-relevant cell subsets and cell type-specific signatures associated with disease activity. Our analysis revealed 18 distinct PBMC subsets, including an IFN-induced transmembrane 3-overexpressing (IFITM3-overexpressing) IFN-activated monocyte subset. We observed an increase in CD4+ T effector memory cells in patients with moderate-high disease activity (DAS28-CRP ≥ 3.2) and a decrease in nonclassical monocytes in patients with low disease activity or remission (DAS28-CRP < 3.2). Pseudobulk analysis by cell type identified 168 differentially expressed genes between RA and matched controls, with a downregulation of proinflammatory genes in the γδ T cell subset, alteration of genes associated with RA predisposition in the IFN-activated subset, and nonclassical monocytes. Additionally, we identified a gene signature associated with moderate-high disease activity, characterized by upregulation of proinflammatory genes such as TNF, JUN, EGR1, IFIT2, MAFB, and G0S2 and downregulation of genes including HLA-DQB1, HLA-DRB5, and TNFSF13B. Notably, cell-cell communication analysis revealed an upregulation of signaling pathways, including VISTA, in both moderate-high and remission-low disease activity contexts. Our findings provide valuable insights into the systemic cellular and molecular mechanisms underlying RA disease activity.
Keywords: Autoimmunity; Immunology; Rheumatology.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

