Trafficking of mitochondrial double-stranded RNA from mitochondria to the cytosol
- PMID: 38955468
- PMCID: PMC11220484
- DOI: 10.26508/lsa.202302396
Trafficking of mitochondrial double-stranded RNA from mitochondria to the cytosol
Abstract
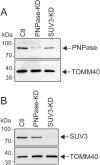
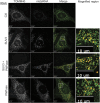
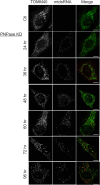
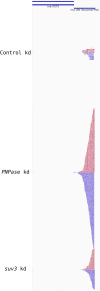
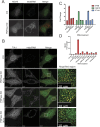
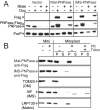
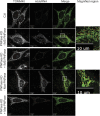
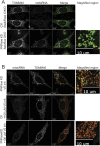
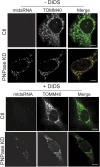
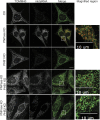
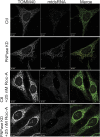
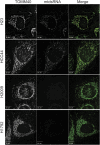
In addition to mitochondrial DNA, mitochondrial double-stranded RNA (mtdsRNA) is exported from mitochondria. However, specific channels for RNA transport have not been demonstrated. Here, we begin to characterize channel candidates for mtdsRNA export from the mitochondrial matrix to the cytosol. Down-regulation of SUV3 resulted in the accumulation of mtdsRNAs in the matrix, whereas down-regulation of PNPase resulted in the export of mtdsRNAs to the cytosol. Targeting experiments show that PNPase functions in both the intermembrane space and matrix. Strand-specific sequencing of the double-stranded RNA confirms the mitochondrial origin. Inhibiting or down-regulating outer membrane proteins VDAC1/2 and BAK/BAX or inner membrane proteins PHB1/2 strongly attenuated the export of mtdsRNAs to the cytosol. The cytosolic mtdsRNAs subsequently localized to large granules containing the stress protein TIA-1 and activated the type 1 interferon stress response pathway. Abundant mtdsRNAs were detected in a subset of non-small-cell lung cancer cell lines that were glycolytic, indicating relevance in cancer biology. Thus, we propose that mtdsRNA is a new damage-associated molecular pattern that is exported from mitochondria in a regulated manner.
© 2024 Krieger et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
