Epigenetic-based differentiation therapy for Acute Myeloid Leukemia
- PMID: 38956053
- PMCID: PMC11219871
- DOI: 10.1038/s41467-024-49784-y
Epigenetic-based differentiation therapy for Acute Myeloid Leukemia
Abstract
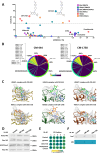
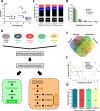
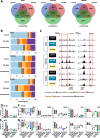
Despite the development of novel therapies for acute myeloid leukemia, outcomes remain poor for most patients, and therapeutic improvements are an urgent unmet need. Although treatment regimens promoting differentiation have succeeded in the treatment of acute promyelocytic leukemia, their role in other acute myeloid leukemia subtypes needs to be explored. Here we identify and characterize two lysine deacetylase inhibitors, CM-444 and CM-1758, exhibiting the capacity to promote myeloid differentiation in all acute myeloid leukemia subtypes at low non-cytotoxic doses, unlike other commercial histone deacetylase inhibitors. Analyzing the acetylome after CM-444 and CM-1758 treatment reveals modulation of non-histone proteins involved in the enhancer-promoter chromatin regulatory complex, including bromodomain proteins. This acetylation is essential for enhancing the expression of key transcription factors directly involved in the differentiation therapy induced by CM-444/CM-1758 in acute myeloid leukemia. In summary, these compounds may represent effective differentiation-based therapeutic agents across acute myeloid leukemia subtypes with a potential mechanism for the treatment of acute myeloid leukemia.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- PI19/01352, PI22/00947/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- PI20/01306, PI23/00488/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- CB16/12/00369/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- 44/2021; 0011-1411-2022-000068/Departamento de Educación, Gobierno de Navarra (Department of Education, Government of Navarra)
- GN2023/11/Departamento de Educación, Gobierno de Navarra (Department of Education, Government of Navarra)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

