Cell signaling and transcriptional regulation of osteoblast lineage commitment, differentiation, bone formation, and homeostasis
- PMID: 38956429
- PMCID: PMC11219878
- DOI: 10.1038/s41421-024-00689-6
Cell signaling and transcriptional regulation of osteoblast lineage commitment, differentiation, bone formation, and homeostasis
Abstract
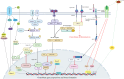
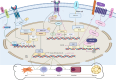
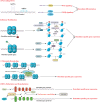
The initiation of osteogenesis primarily occurs as mesenchymal stem cells undergo differentiation into osteoblasts. This differentiation process plays a crucial role in bone formation and homeostasis and is regulated by two intricate processes: cell signal transduction and transcriptional gene expression. Various essential cell signaling pathways, including Wnt, BMP, TGF-β, Hedgehog, PTH, FGF, Ephrin, Notch, Hippo, and Piezo1/2, play a critical role in facilitating osteoblast differentiation, bone formation, and bone homeostasis. Key transcriptional factors in this differentiation process include Runx2, Cbfβ, Runx1, Osterix, ATF4, SATB2, and TAZ/YAP. Furthermore, a diverse array of epigenetic factors also plays critical roles in osteoblast differentiation, bone formation, and homeostasis at the transcriptional level. This review provides an overview of the latest developments and current comprehension concerning the pathways of cell signaling, regulation of hormones, and transcriptional regulation of genes involved in the commitment and differentiation of osteoblast lineage, as well as in bone formation and maintenance of homeostasis. The paper also reviews epigenetic regulation of osteoblast differentiation via mechanisms, such as histone and DNA modifications. Additionally, we summarize the latest developments in osteoblast biology spurred by recent advancements in various modern technologies and bioinformatics. By synthesizing these insights into a comprehensive understanding of osteoblast differentiation, this review provides further clarification of the mechanisms underlying osteoblast lineage commitment, differentiation, and bone formation, and highlights potential new therapeutic applications for the treatment of bone diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
Grants and funding
- AG056438/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01 DE023813/DE/NIDCR NIH HHS/United States
- AR075735/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- DE028264/U.S. Department of Health & Human Services | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- R01 AG056438/AG/NIA NIH HHS/United States
- R01 AR070135/AR/NIAMS NIH HHS/United States
- AR074954/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- DE023813/U.S. Department of Health & Human Services | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- R01 AR075735/AR/NIAMS NIH HHS/United States
- R01 DE028264/DE/NIDCR NIH HHS/United States
- AR070135/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- R01 AR074954/AR/NIAMS NIH HHS/United States
- R56 AG056438/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources

