Exploring selection signatures in the divergence and evolution of lipid droplet (LD) associated genes in major oilseed crops
- PMID: 38956471
- PMCID: PMC11218257
- DOI: 10.1186/s12864-024-10527-4
Exploring selection signatures in the divergence and evolution of lipid droplet (LD) associated genes in major oilseed crops
Abstract
Background: Oil bodies or lipid droplets (LDs) in the cytosol are the subcellular storage compartments of seeds and the sites of lipid metabolism providing energy to the germinating seeds. Major LD-associated proteins are lipoxygenases, phospholipaseD, oleosins, TAG-lipases, steroleosins, caleosins and SEIPINs; involved in facilitating germination and enhancing peroxidation resulting in off-flavours. However, how natural selection is balancing contradictory processes in lipid-rich seeds remains evasive. The present study was aimed at the prediction of selection signatures among orthologous clades in major oilseeds and the correlation of selection effect with gene expression.
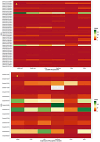
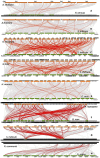
Results: The LD-associated genes from the major oil-bearing crops were analyzed to predict natural selection signatures in phylogenetically close-knit ortholog clusters to understand adaptive evolution. Positive selection was the major force driving the evolution and diversification of orthologs in a lineage-specific manner. Significant positive selection effects were found in 94 genes particularly in oleosin and TAG-lipases, purifying with excess of non-synonymous substitution in 44 genes while 35 genes were neutral to selection effects. No significant selection impact was noticed in Brassicaceae as against LOX genes of oil palm. A heavy load of deleterious mutations affecting selection signatures was detected in T-lineage oleosins and LOX genes of Arachis hypogaea. The T-lineage oleosin genes were involved in mainly anther, tapetum and anther wall morphogenesis. In Ricinus communis and Sesamum indicum > 85% of PLD genes were under selection whereas selection pressures were low in Brassica juncea and Helianthus annuus. Steroleosin, caleosin and SEIPINs with large roles in lipid droplet organization expressed mostly in seeds and were under considerable positive selection pressures. Expression divergence was evident among paralogs and homeologs with one gene attaining functional superiority compared to the other. The LOX gene Glyma.13g347500 associated with off-flavor was not expressed during germination, rather its paralog Glyma.13g347600 showed expression in Glycine max. PLD-α genes were expressed on all the tissues except the seed,δ genes in seed and meristem while β and γ genes expressed in the leaf.
Conclusions: The genes involved in seed germination and lipid metabolism were under strong positive selection, although species differences were discernable. The present study identifies suitable candidate genes enhancing seed oil content and germination wherein directional selection can become more fruitful.
Keywords: Germination; Lipid droplet; Lipoxygenase; Natural selection; Oleosin; Phospholipase D; Rancidity; TAG lipase.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Fadda A, Sanna D, Sakar EH, Gharby S, Mulas M, Medda S, Yesilcubuk NS, Karaca AC, Gozukirmizi CK, Lucarini M, Lombardi-Boccia G. Innovative and sustainable technologies to enhance the oxidative stability of vegetable oils. Sustainability. 2022;14(2):849. doi: 10.3390/su14020849. - DOI
-
- Le Moigne D, Guéguen N, Salvaing J. Lipid droplets in plants: more than a simple fat storage. Adv Bot Res. 2022;101:191–223. doi: 10.1016/bs.abr.2021.07.004. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

