Streamlining heterologous expression of top carbonic anhydrases in Escherichia coli: bioinformatic and experimental approaches
- PMID: 38956607
- PMCID: PMC11218372
- DOI: 10.1186/s12934-024-02463-5
Streamlining heterologous expression of top carbonic anhydrases in Escherichia coli: bioinformatic and experimental approaches
Abstract
Background: Carbonic anhydrase (CA) enzymes facilitate the reversible hydration of CO2 to bicarbonate ions and protons. Identifying efficient and robust CAs and expressing them in model host cells, such as Escherichia coli, enables more efficient engineering of these enzymes for industrial CO2 capture. However, expression of CAs in E. coli is challenging due to the possible formation of insoluble protein aggregates, or inclusion bodies. This makes the production of soluble and active CA protein a prerequisite for downstream applications.
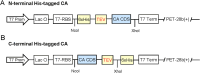
Results: In this study, we streamlined the process of CA expression by selecting seven top CA candidates and used two bioinformatic tools to predict their solubility for expression in E. coli. The prediction results place these enzymes in two categories: low and high solubility. Our expression of high solubility score CAs (namely CA5-SspCA, CA6-SazCAtrunc, CA7-PabCA and CA8-PhoCA) led to significantly higher protein yields (5 to 75 mg purified protein per liter) in flask cultures, indicating a strong correlation between the solubility prediction score and protein expression yields. Furthermore, phylogenetic tree analysis demonstrated CA class-specific clustering patterns for protein solubility and production yields. Unexpectedly, we also found that the unique N-terminal, 11-amino acid segment found after the signal sequence (not present in its homologs), was essential for CA6-SazCA activity.
Conclusions: Overall, this work demonstrated that protein solubility prediction, phylogenetic tree analysis, and experimental validation are potent tools for identifying top CA candidates and then producing soluble, active forms of these enzymes in E. coli. The comprehensive approaches we report here should be extendable to the expression of other heterogeneous proteins in E. coli.
Keywords: Escherichia coli; Carbonic anhydrase; Phylogenetic tree analysis; Protein expression; Protein solubility prediction.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- WBS 5.1.3.103/U.S. Department of Energy Office of Energy Efficiency and Renewable Energy (EERE) Bioenergy Technologies Office (BETO)
- WBS 5.1.3.103/U.S. Department of Energy Office of Energy Efficiency and Renewable Energy (EERE) Bioenergy Technologies Office (BETO)
- WBS 5.1.3.103/U.S. Department of Energy Office of Energy Efficiency and Renewable Energy (EERE) Bioenergy Technologies Office (BETO)
- WBS 5.1.3.103/U.S. Department of Energy Office of Energy Efficiency and Renewable Energy (EERE) Bioenergy Technologies Office (BETO)
- WBS 5.1.3.103/U.S. Department of Energy Office of Energy Efficiency and Renewable Energy (EERE) Bioenergy Technologies Office (BETO)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

