An expedited screening platform for the discovery of anti-ageing compounds in vitro and in vivo
- PMID: 38956711
- PMCID: PMC11218148
- DOI: 10.1186/s13073-024-01349-w
An expedited screening platform for the discovery of anti-ageing compounds in vitro and in vivo
Abstract
Background: Restraining or slowing ageing hallmarks at the cellular level have been proposed as a route to increased organismal lifespan and healthspan. Consequently, there is great interest in anti-ageing drug discovery. However, this currently requires laborious and lengthy longevity analysis. Here, we present a novel screening readout for the expedited discovery of compounds that restrain ageing of cell populations in vitro and enable extension of in vivo lifespan.
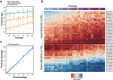
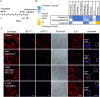
Methods: Using Illumina methylation arrays, we monitored DNA methylation changes accompanying long-term passaging of adult primary human cells in culture. This enabled us to develop, test, and validate the CellPopAge Clock, an epigenetic clock with underlying algorithm, unique among existing epigenetic clocks for its design to detect anti-ageing compounds in vitro. Additionally, we measured markers of senescence and performed longevity experiments in vivo in Drosophila, to further validate our approach to discover novel anti-ageing compounds. Finally, we bench mark our epigenetic clock with other available epigenetic clocks to consolidate its usefulness and specialisation for primary cells in culture.
Results: We developed a novel epigenetic clock, the CellPopAge Clock, to accurately monitor the age of a population of adult human primary cells. We find that the CellPopAge Clock can detect decelerated passage-based ageing of human primary cells treated with rapamycin or trametinib, well-established longevity drugs. We then utilise the CellPopAge Clock as a screening tool for the identification of compounds which decelerate ageing of cell populations, uncovering novel anti-ageing drugs, torin2 and dactolisib (BEZ-235). We demonstrate that delayed epigenetic ageing in human primary cells treated with anti-ageing compounds is accompanied by a reduction in senescence and ageing biomarkers. Finally, we extend our screening platform in vivo by taking advantage of a specially formulated holidic medium for increased drug bioavailability in Drosophila. We show that the novel anti-ageing drugs, torin2 and dactolisib (BEZ-235), increase longevity in vivo.
Conclusions: Our method expands the scope of CpG methylation profiling to accurately and rapidly detecting anti-ageing potential of drugs using human cells in vitro, and in vivo, providing a novel accelerated discovery platform to test sought after anti-ageing compounds and geroprotectors.
Keywords: Ageing; CellPopAge epigenetic Clock; CpG methylation; Drug discovery; Rapamycin; Senescence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

