mTORC1 Signaling in Brain Endothelial Progenitors Contributes to CCM Pathogenesis
- PMID: 38957991
- PMCID: PMC11293987
- DOI: 10.1161/CIRCRESAHA.123.324015
mTORC1 Signaling in Brain Endothelial Progenitors Contributes to CCM Pathogenesis
Erratum in
-
Correction to: mTORC1 Signaling in Brain Endothelial Progenitors Contributes to CCM Pathogenesis.Circ Res. 2025 Jun 20;137(1):e16-e17. doi: 10.1161/RES.0000000000000718. Epub 2025 Jun 19. Circ Res. 2025. PMID: 40536942 No abstract available.
Abstract
Background: Cerebral vascular malformations (CCMs) are primarily found within the brain, where they result in increased risk for stroke, seizures, and focal neurological deficits. The unique feature of the brain vasculature is the blood-brain barrier formed by the brain neurovascular unit. Recent studies suggest that loss of CCM genes causes disruptions of blood-brain barrier integrity as the inciting events for CCM development. CCM lesions are proposed to be initially derived from a single clonal expansion of a subset of angiogenic venous capillary endothelial cells (ECs) and respective resident endothelial progenitor cells (EPCs). However, the critical signaling events in the subclass of brain ECs/EPCs for CCM lesion initiation and progression are unclear.
Methods: Brain EC-specific CCM3-deficient (Pdcd10BECKO) mice were generated by crossing Pdcd10fl/fl mice with Mfsd2a-CreERT2 mice. Single-cell RNA-sequencing analyses were performed by the chromium single-cell platform (10× genomics). Cell clusters were annotated into EC subtypes based on visual inspection and GO analyses. Cerebral vessels were visualized by 2-photon in vivo imaging and tissue immunofluorescence analyses. Regulation of mTOR (mechanistic target of rapamycin) signaling by CCM3 and Cav1 (caveolin-1) was performed by cell biology and biochemical approaches.
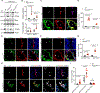
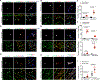
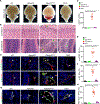
Results: Single-cell RNA-sequencing analyses from P10 Pdcd10BECKO mice harboring visible CCM lesions identified upregulated CCM lesion signature and mitotic EC clusters but decreased blood-brain barrier-associated EC clusters. However, a unique EPC cluster with high expression levels of stem cell markers enriched with mTOR signaling was identified from early stages of the P6 Pdcd10BECKO brain. Indeed, mTOR signaling was upregulated in both mouse and human CCM lesions. Genetic deficiency of Raptor (regulatory-associated protein of mTOR), but not of Rictor (rapamycin-insensitive companion of mTOR), prevented CCM lesion formation in the Pdcd10BECKO model. Importantly, the mTORC1 (mTOR complex 1) pharmacological inhibitor rapamycin suppressed EPC proliferation and ameliorated CCM pathogenesis in Pdcd10BECKO mice. Mechanistic studies suggested that Cav1/caveolae increased in CCM3-depleted EPC-mediated intracellular trafficking and complex formation of the mTORC1 signaling proteins.
Conclusions: CCM3 is critical for maintaining blood-brain barrier integrity and CCM3 loss-induced mTORC1 signaling in brain EPCs initiates and facilitates CCM pathogenesis.
Keywords: TOR serine-threonine kinases; blood–brain barrier; caveolae; endocytosis; hemangioma, cavernous, central nervous system; mechanistic target of rapamycin complex 1; single-cell gene expression analysis.
Conflict of interest statement
None.
Figures





References
-
- Otten P, Pizzolato GP, Rilliet B and Berney J. [131 cases of cavernous angioma (cavernomas) of the CNS, discovered by retrospective analysis of 24,535 autopsies]. Neurochirurgie. 1989;35:82–3, 128–31. - PubMed
-
- Rigamonti D, Hadley MN, Drayer BP, Johnson PC, Hoenig-Rigamonti K, Knight JT and Spetzler RF. Cerebral cavernous malformations. Incidence and familial occurrence. N Engl J Med. 1988;319:343–7. - PubMed
-
- Cavalcanti DD, Kalani MY, Martirosyan NL, Eales J, Spetzler RF and Preul MC. Cerebral cavernous malformations: from genes to proteins to disease. Journal of neurosurgery. 2012;116:122–32. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

