Molecular epidemiology of enteroviruses from Guatemalan wastewater isolated from human lung fibroblasts
- PMID: 38959255
- PMCID: PMC11221682
- DOI: 10.1371/journal.pone.0305108
Molecular epidemiology of enteroviruses from Guatemalan wastewater isolated from human lung fibroblasts
Abstract
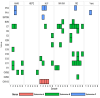
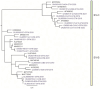
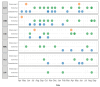
The Global Specialized Polio Laboratory at CDC supports the Global Poliovirus Laboratory Network with environmental surveillance (ES) to detect the presence of vaccine strain polioviruses, vaccine-derived polioviruses, and wild polioviruses in high-risk countries. Environmental sampling provides valuable supplementary information, particularly in areas with gaps in surveillance of acute flaccid paralysis (AFP) mainly in children less than 15 years. In collaboration with Guatemala's National Health Laboratory (Laboratorio Nacional de Salud Guatemala), monthly sewage collections allowed screening enterovirus (EV) presence without incurring additional costs for sample collection, transport, or concentration. Murine recombinant fibroblast L-cells (L20B) and human rhabdomyosarcoma (RD) cells are used for the isolation of polioviruses following a standard detection algorithm. Though non-polio-Enteroviruses (NPEV) can be isolated, the algorithm is optimized for the detection of polioviruses. To explore if other EV's are present in sewage not found through standard methods, five additional cell lines were piloted in a small-scale experiment, and next-generation sequencing (NGS) was used for the identification of any EV types. Human lung fibroblast cells (HLF) were selected based on their ability to isolate EV-A genus. Sewage concentrates collected between 2020-2021 were isolated in HLF cells and any cytopathic effect positive isolates used for NGS. A large variety of EVs, including echoviruses 1, 3, 6, 7, 11, 13, 18, 19, 25, 29; coxsackievirus A13, B2, and B5, EV-C99, EVB, and polioviruses (Sabin 1 and 3) were identified through genomic typing in NGS. When the EV genotypes were compared by phylogenetic analysis, it showed many EV's were genomically like viruses previously isolated from ES collected in Haiti. Enterovirus occurrence did not follow a seasonality, but more diverse EV types were found in ES collection sites with lower populations. Using the additional cell line in the existing poliovirus ES algorithm may add value by providing data about EV circulation, without additional sample collection or processing. Next-generation sequencing closed gaps in knowledge providing molecular epidemiological information on multiple EV types and full genome sequences of EVs present in wastewater in Guatemala.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Craighead JE. CHAPTER 1—Enteroviruses. In: Craighead JE, editor. Pathology and Pathogenesis of Human Viral Disease. San Diego: Academic Press; 2000. p. 1–28.
-
- Pogka V, Labropoulou S, Emmanouil M, Voulgari-Kokota A, Vernardaki A, Georgakopoulou T, et al.. Laboratory Surveillance of Polio and Other Enteroviruses in High-Risk Populations and Environmental Samples. Appl Environ Microbiol. 2017;83(5). Epub 20170215. doi: 10.1128/AEM.02872-16 . - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

