Optimizing the timing of an end-of-outbreak declaration: Ebola virus disease in the Democratic Republic of the Congo
- PMID: 38959306
- PMCID: PMC11221504
- DOI: 10.1126/sciadv.ado7576
Optimizing the timing of an end-of-outbreak declaration: Ebola virus disease in the Democratic Republic of the Congo
Abstract
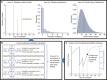
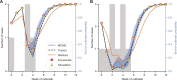
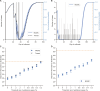
Following the apparent final case in an Ebola virus disease (EVD) outbreak, the decision to declare the outbreak over must balance societal benefits of relaxing interventions against the risk of resurgence. Estimates of the end-of-outbreak probability (the probability that no future cases will occur) provide quantitative evidence that can inform the timing of an end-of-outbreak declaration. An existing modeling approach for estimating the end-of-outbreak probability requires comprehensive contact tracing data describing who infected whom to be available, but such data are often unavailable or incomplete during outbreaks. Here, we develop a Markov chain Monte Carlo-based approach that extends the previous method and does not require contact tracing data. Considering data from two EVD outbreaks in the Democratic Republic of the Congo, we find that data describing who infected whom are not required to resolve uncertainty about when to declare an outbreak over.
Figures
Similar articles
-
"We have to amplify what we saw at EBOVAC" - Assessing participant perceptions, attitudes, and acceptability of an ancillary care policy in an Ebola vaccine trial in the Democratic Republic of the Congo: A mixed methods study.PLoS One. 2025 Jun 24;20(6):e0325435. doi: 10.1371/journal.pone.0325435. eCollection 2025. PLoS One. 2025. PMID: 40554528 Free PMC article.
-
A systematic review of Ebola virus disease outbreaks and an analysis of the efficacy and safety of newer drugs approved for the treatment of Ebola virus disease by the US Food and Drug Administration from 2016 to 2020.J Infect Public Health. 2022 Mar;15(3):285-292. doi: 10.1016/j.jiph.2022.01.005. Epub 2022 Jan 21. J Infect Public Health. 2022. PMID: 35085865
-
The Ebola virus - going beyond the bleeding edge.J Med Microbiol. 2025 Jul;74(7):001998. doi: 10.1099/jmm.0.001998. J Med Microbiol. 2025. PMID: 40607496 Free PMC article. Review.
-
2020 Ebola virus disease outbreak in Équateur Province, Democratic Republic of the Congo: a retrospective genomic characterisation.Lancet Microbe. 2024 Feb;5(2):e109-e118. doi: 10.1016/S2666-5247(23)00259-8. Epub 2024 Jan 24. Lancet Microbe. 2024. PMID: 38278165 Free PMC article.
-
Operational and Workforce Capacity Improvements for Supporting Public Health Emergency Management: Lessons Learned for Preparing for and Responding to 2014-2022 Ebola Outbreaks in Africa.Health Promot Pract. 2025 Sep;26(5):985-992. doi: 10.1177/15248399241294236. Epub 2024 Nov 19. Health Promot Pract. 2025. PMID: 39560125
Cited by
-
Estimation of end-of-outbreak probabilities in the presence of delayed and incomplete case reporting.Proc Biol Sci. 2025 Jan;292(2039):20242825. doi: 10.1098/rspb.2024.2825. Epub 2025 Jan 29. Proc Biol Sci. 2025. PMID: 39876727 Free PMC article.
-
Real-time inference of the end of an outbreak: Temporally aggregated disease incidence data and under-reporting.Infect Dis Model. 2025 Apr 1;10(3):935-945. doi: 10.1016/j.idm.2025.03.009. eCollection 2025 Sep. Infect Dis Model. 2025. PMID: 40475698 Free PMC article.
-
Reducing transmission in multiple settings is required to eliminate the risk of major Ebola outbreaks: a mathematical modelling study.J R Soc Interface. 2025 Mar;22(224):20240765. doi: 10.1098/rsif.2024.0765. Epub 2025 Mar 19. J R Soc Interface. 2025. PMID: 40101777 Free PMC article.
References
-
- World Health Organization, Ebola virus disease (2021); https://who.int/news-room/fact-sheets/detail/ebola-virus-disease.
-
- World Health Organization, WHO recommended criteria for declaring the end of the Ebola Virus Disease Outbreak (2020); https://who.int/publications/m/item/who-recommended-criteria-for-declari....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

