CSF Proteomics in Patients With Progressive Supranuclear Palsy
- PMID: 38959435
- PMCID: PMC11226322
- DOI: 10.1212/WNL.0000000000209585
CSF Proteomics in Patients With Progressive Supranuclear Palsy
Abstract
Background and objectives: Identification of fluid biomarkers for progressive supranuclear palsy (PSP) is critical to enhance therapeutic development. We implemented unbiased DNA aptamer (SOMAmer) proteomics to identify novel CSF PSP biomarkers.
Methods: This is a cross-sectional study in original (18 clinically diagnosed PSP-Richardson syndrome [PSP-RS], 28 cognitively healthy controls]), validation (23 PSP-RS, 26 healthy controls), and neuropathology-confirmed (21 PSP, 52 non-PSP frontotemporal lobar degeneration) cohorts. Participants were recruited through the University of California, San Francisco, and the 4-Repeat Neuroimaging Initiative. The original and neuropathology cohorts were analyzed with the SomaScan platform version 3.0 (5026-plex) and the validation cohort with version 4.1 (7595-plex). Clinical severity was measured with the PSP Rating Scale (PSPRS). CSF proteomic data were analyzed to identify differentially expressed targets, implicated biological pathways using enrichment and weighted consensus gene coexpression analyses, diagnostic value of top targets with receiver-operating characteristic curves, and associations with disease severity with linear regressions.
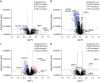
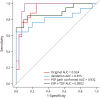
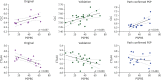
Results: A total of 136 participants were included (median age 70.6 ± 8 years, 68 [50%] women). One hundred fifty-five of 5,026 (3.1%), 959 of 7,595 (12.6%), and 321 of 5,026 (6.3%) SOMAmers were differentially expressed in PSP compared with controls in original, validation, and neuropathology-confirmed cohorts, with most of the SOMAmers showing reduced signal (83.1%, 95.1%, and 73.2%, respectively). Three coexpression modules were associated with PSP across cohorts: (1) synaptic function/JAK-STAT (β = -0.044, corrected p = 0.002), (2) vesicle cytoskeletal trafficking (β = 0.039, p = 0.007), and (3) cytokine-cytokine receptor interaction (β = -0.032, p = 0.035) pathways. Axon guidance was the top dysregulated pathway in PSP in original (strength = 1.71, p < 0.001), validation (strength = 0.84, p < 0.001), and neuropathology-confirmed (strength = 0.78, p < 0.001) cohorts. A panel of axon guidance pathway proteins discriminated between PSP and controls in original (area under the curve [AUC] = 0.924), validation (AUC = 0.815), and neuropathology-confirmed (AUC = 0.932) cohorts. Two inflammatory proteins, galectin-10 and cytotoxic T lymphocyte-associated protein-4, correlated with PSPRS scores across cohorts.
Discussion: Axon guidance pathway proteins and several other molecular pathways are downregulated in PSP, compared with controls. Proteins in these pathways may be useful targets for biomarker or therapeutic development.
Conflict of interest statement
L. Mitic is supported by the Bluefield Project to Cure FTD. L. Vandevrede is supported by K23AG073514 and grants from the Alzheimer's Association and American Academy of Neurology, site PI for clinical trials sponsored by Biogen, consulted for Retrotope. M. Grossman is deceased, to the best of our knowledge, he had no relevant disclosures. A. Pantelyat is supported by NIH/NIA K23AG059891, NIH/National Institute of Neurological Disorders and Stroke U01NS102035; NIH/NIA R01AG038791; Scientific Advisory Board, MedRhythms, Inc. J.C. Rojas NIA/NIH K23AG59888 is site principal investigator for Clinical trials sponsored by Eisai and Eli-Lilly. Go to
Figures

References
-
- Greene P. Parkinson-plus syndromes. In: Rowland LP, Pedley TA, eds. Merrit's Neurology. 12th ed. Lippincott Williams & Wilkins; 2010:770-774.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
