Histone H3K18 & H3K23 acetylation directs establishment of MLL-mediated H3K4 methylation
- PMID: 38960040
- PMCID: PMC11338103
- DOI: 10.1016/j.jbc.2024.107527
Histone H3K18 & H3K23 acetylation directs establishment of MLL-mediated H3K4 methylation
Abstract
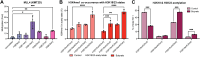
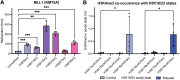
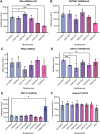
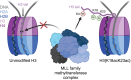
In an unmodified state, positively charged histone N-terminal tails engage nucleosomal DNA in a manner which restricts access to not only the underlying DNA but also key tail residues subject to binding and/or modification. Charge-neutralizing modifications, such as histone acetylation, serve to disrupt this DNA-tail interaction, facilitating access to such residues. We previously showed that a polyacetylation-mediated chromatin "switch" governs the read-write capability of H3K4me3 by the MLL1 methyltransferase complex. Here, we discern the relative contributions of site-specific acetylation states along the H3 tail and extend our interrogation to other chromatin modifiers. We show that the contributions of H3 tail acetylation to H3K4 methylation by MLL1 are highly variable, with H3K18 and H3K23 acetylation exhibiting robust stimulatory effects and that this extends to the related H3K4 methyltransferase complex, MLL4. We show that H3K4me1 and H3K4me3 are found preferentially co-enriched with H3 N-terminal tail proteoforms bearing dual H3K18 and H3K23 acetylation (H3{K18acK23ac}). We further show that this effect is specific to H3K4 methylation, while methyltransferases targeting other H3 tail residues (H3K9, H3K27, & H3K36), a methyltransferase targeting the nucleosome core (H3K79), and a kinase targeting a residue directly adjacent to H3K4 (H3T3) are insensitive to tail acetylation. Together, these findings indicate a unique and robust stimulation of H3K4 methylation by H3K18 and H3K23 acetylation and provide key insight into why H3K4 methylation is often associated with histone acetylation in the context of active gene expression.
Keywords: chromatin; epigenetics; histone acetylation; histone methylation; nucleosome.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest EpiCypher is a commercial developer and supplier of fully defined semi-synthetic nucleosomes as used in this study. N. N. R., B. G., and B. D. S. own shares in EpiCypher with BDS also a board member of the same. All other authors declare that they have no other conflicts of interest with the contents of this article.
Figures
Update of
-
H3K18 & H3K23 acetylation directs establishment of MLL-mediated H3K4 methylation.bioRxiv [Preprint]. 2024 May 14:2024.05.13.590588. doi: 10.1101/2024.05.13.590588. bioRxiv. 2024. Update in: J Biol Chem. 2024 Aug;300(8):107527. doi: 10.1016/j.jbc.2024.107527. PMID: 38798640 Free PMC article. Updated. Preprint.
Similar articles
-
H3K18 & H3K23 acetylation directs establishment of MLL-mediated H3K4 methylation.bioRxiv [Preprint]. 2024 May 14:2024.05.13.590588. doi: 10.1101/2024.05.13.590588. bioRxiv. 2024. Update in: J Biol Chem. 2024 Aug;300(8):107527. doi: 10.1016/j.jbc.2024.107527. PMID: 38798640 Free PMC article. Updated. Preprint.
-
Structural basis of nucleosome recognition and modification by MLL methyltransferases.Nature. 2019 Sep;573(7774):445-449. doi: 10.1038/s41586-019-1528-1. Epub 2019 Sep 4. Nature. 2019. PMID: 31485071
-
An acetylation-mediated chromatin switch governs H3K4 methylation read-write capability.Elife. 2023 May 19;12:e82596. doi: 10.7554/eLife.82596. Elife. 2023. PMID: 37204295 Free PMC article.
-
Why are so many MLL lysine methyltransferases required for normal mammalian development?Cell Mol Life Sci. 2019 Aug;76(15):2885-2898. doi: 10.1007/s00018-019-03143-z. Epub 2019 May 16. Cell Mol Life Sci. 2019. PMID: 31098676 Free PMC article. Review.
-
Insights on the regulation of the MLL/SET1 family histone methyltransferases.Biochim Biophys Acta Gene Regul Mech. 2020 Jul;1863(7):194561. doi: 10.1016/j.bbagrm.2020.194561. Epub 2020 Apr 15. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32304759 Free PMC article. Review.
Cited by
-
The epigenetic circle: feedback loops in the maintenance of cellular memory.Epigenetics Chromatin. 2025 Aug 20;18(1):56. doi: 10.1186/s13072-025-00621-6. Epigenetics Chromatin. 2025. PMID: 40830515 Free PMC article. Review.
-
Normalized and Directional Interplay Scoring for the Interrogation of Proteoform Data.bioRxiv [Preprint]. 2024 Nov 18:2024.11.18.624157. doi: 10.1101/2024.11.18.624157. bioRxiv. 2024. Update in: J Proteome Res. 2025 Apr 04;24(4):1765-1777. doi: 10.1021/acs.jproteome.4c00877. PMID: 39605462 Free PMC article. Updated. Preprint.
-
SETD2 suppresses tumorigenesis in a KRASG12C-driven lung cancer model and its catalytic activity is regulated by histone acetylation.bioRxiv [Preprint]. 2025 Jul 14:2025.05.16.654513. doi: 10.1101/2025.05.16.654513. bioRxiv. 2025. PMID: 40462961 Free PMC article. Preprint.
-
Capillary Electrophoresis-Mass Spectrometry for Top-Down Proteomics.Annu Rev Anal Chem (Palo Alto Calif). 2025 May;18(1):125-147. doi: 10.1146/annurev-anchem-071124-092242. Epub 2025 Jan 23. Annu Rev Anal Chem (Palo Alto Calif). 2025. PMID: 39847747 Review.
References
-
- Strahl B.D., David Allis C. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Lo W.-S., Trievel R.C., Rojas J.R., Duggan L., Hsu J.-Y., David Allis C., et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 2000;7:917–926. - PubMed
-
- Hassan A.H., Prochasson P., Neely K.E., Galasinski S.C., Chandy M., Carrozza M.J., et al. Function and Selectivity of Bromodomains in Anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG085751/AG/NIA NIH HHS/United States
- T32 GM135128/GM/NIGMS NIH HHS/United States
- R35 GM152103/GM/NIGMS NIH HHS/United States
- R01 AG074540/AG/NIA NIH HHS/United States
- P01 AG066606/AG/NIA NIH HHS/United States
- T32 CA217824/CA/NCI NIH HHS/United States
- R01 CA193235/CA/NCI NIH HHS/United States
- R01 NS136375/NS/NINDS NIH HHS/United States
- R01 GM139295/GM/NIGMS NIH HHS/United States
- F32 CA139893/CA/NCI NIH HHS/United States
- R35 GM124764/GM/NIGMS NIH HHS/United States
- R35 GM126900/GM/NIGMS NIH HHS/United States
- R01 CA276663/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

