Unravelling reference bias in ancient DNA datasets
- PMID: 38960861
- PMCID: PMC11254355
- DOI: 10.1093/bioinformatics/btae436
Unravelling reference bias in ancient DNA datasets
Abstract
Motivation: The alignment of sequencing reads is a critical step in the characterization of ancient genomes. However, reference bias and spurious mappings pose a significant challenge, particularly as cutting-edge wet lab methods generate datasets that push the boundaries of alignment tools. Reference bias occurs when reference alleles are favoured over alternative alleles during mapping, whereas spurious mappings stem from either contamination or when endogenous reads fail to align to their correct position. Previous work has shown that these phenomena are correlated with read length but a more thorough investigation of reference bias and spurious mappings for ancient DNA has been lacking. Here, we use a range of empirical and simulated palaeogenomic datasets to investigate the impacts of mapping tools, quality thresholds, and reference genome on mismatch rates across read lengths.
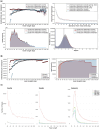
Results: For these analyses, we introduce AMBER, a new bioinformatics tool for assessing the quality of ancient DNA mapping directly from BAM-files and informing on reference bias, read length cut-offs and reference selection. AMBER rapidly and simultaneously computes the sequence read mapping bias in the form of the mismatch rates per read length, cytosine deamination profiles at both CpG and non-CpG sites, fragment length distributions, and genomic breadth and depth of coverage. Using AMBER, we find that mapping algorithms and quality threshold choices dictate reference bias and rates of spurious alignment at different read lengths in a predictable manner, suggesting that optimized mapping parameters for each read length will be a key step in alleviating reference bias and spurious mappings.
Availability and implementation: AMBER is available for noncommercial use on GitHub (https://github.com/tvandervalk/AMBER.git). Scripts used to generate and analyse simulated datasets are available on Github (https://github.com/sdolenz/refbias_scripts).
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
-
- Fernandez-Guerra A, Borrel G, Delmont TO. et al. A 2-million-year-old microbial and viral communities from the Kap København Formation in North Greenland. bioRxiv, 10.1101/2023.06.10.544454, 2023, preprint: not peer reviewed. - DOI

