miR-708-3p targetedly regulates LSD1 to promote osteoblast differentiation of hPDLSCs in periodontitis
- PMID: 38961043
- PMCID: PMC11717837
- DOI: 10.1007/s10266-024-00963-9
miR-708-3p targetedly regulates LSD1 to promote osteoblast differentiation of hPDLSCs in periodontitis
Erratum in
-
Correction: miR-708-3p targetedly regulates LSD1 to promote osteoblast differentiation of hPDLSCs in periodontitis.Odontology. 2025 Jan;113(1):231. doi: 10.1007/s10266-024-00983-5. Odontology. 2025. PMID: 39060897 Free PMC article. No abstract available.
Abstract
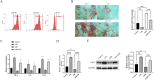
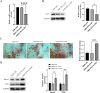
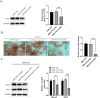
Periodontitis (PD) is a multifactorial inflammatory disease associated with periodontopathic bacteria. Lysine-specific demethylase 1 (LSD1), a type of histone demethylase, has been implicated in the modulation of the inflammatory response process in oral diseases by binding to miRNA targets. This study investigates the molecular mechanisms by which miRNA binds to LSD1 and its subsequent effect on osteogenic differentiation. First, human periodontal ligament stem cells (hPDLSCs) were isolated, cultured, and characterized. These cells were then subjected to lipopolysaccharide (LPS) treatment to induce inflammation, after which osteogenic differentiation was initiated. qPCR and western blot were employed to monitor changes in LSD1 expression. Subsequently, LSD1 was silenced in hPDLSCs to evaluate its impact on osteogenic differentiation. Through bioinformatics and dual luciferase reporter assay, miR-708-3p was predicted and confirmed as a target miRNA of LSD1. Subsequently, miR-708-3p expression was assessed, and its role in hPDLSCs in PD was evaluated through overexpression. Using chromatin immunoprecipitation (ChIP) and western blot assay, we explored the potential regulation of osterix (OSX) transcription by miR-708-3p and LSD1 via di-methylated H3K4 (H3K4me2). Finally, we investigated the role of OSX in hPDLSCs. Following LPS treatment of hPDLSCs, the expression of LSD1 increased, but this trend was reversed upon the induction of osteogenic differentiation. Silencing LSD1 strengthened the osteogenic differentiation of hPDLSCs. miR-708-3p was found to directly bind to and negatively regulate LSD1, leading to the repression of OSX transcription through demethylation of H3K4me2. Moreover, overexpression of miR-708-3p was found to promote hPDLSCs osteogenic differentiation in inflammatory microenvironment. However, the protective effect was partially attenuated by reduced expression of OSX. Our findings indicate that miR-708-3p targetedly regulates LSD1 to enhance OSX transcription via H3K4me2 methylation, ultimately promoting hPDLSCs osteogenic differentiation.
Keywords: Inflammation; LSD1; Osteogenesis; Periodontitis; miR-708-3p.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare that they have no conflict of interest. Ethical standards: The research was carried out following the principles of the Declaration of Helsinki and was approved by the Ethics Committee of Foshan Stomatological Hospital protocol. Signed informed consent was also obtained from all participants or guardians. Consent for publication: The work described has not been published previously.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

