ezSingleCell: an integrated one-stop single-cell and spatial omics analysis platform for bench scientists
- PMID: 38961061
- PMCID: PMC11222513
- DOI: 10.1038/s41467-024-48188-2
ezSingleCell: an integrated one-stop single-cell and spatial omics analysis platform for bench scientists
Abstract
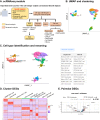
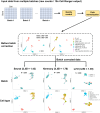
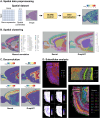
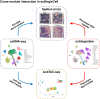
ezSingleCell is an interactive and easy-to-use application for analysing various single-cell and spatial omics data types without requiring prior programing knowledge. It combines the best-performing publicly available methods for in-depth data analysis, integration, and interactive data visualization. ezSingleCell consists of five modules, each designed to be a comprehensive workflow for one data type or task. In addition, ezSingleCell allows crosstalk between different modules within a unified interface. Acceptable input data can be in a variety of formats while the output consists of publication ready figures and tables. In-depth manuals and video tutorials are available to guide users on the analysis workflows and parameter adjustments to suit their study aims. ezSingleCell's streamlined interface can analyse a standard scRNA-seq dataset of 3000 cells in less than five minutes. ezSingleCell is available in two forms: an installation-free web application ( https://immunesinglecell.org/ezsc/ ) or a software package with a shinyApp interface ( https://github.com/JinmiaoChenLab/ezSingleCell2 ) for offline analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Grants and funding
- C211118015/Agency for Science, Technology and Research (A*STAR)
- NMRC-OFIRG18nov-2013/MOH | National Medical Research Council (NMRC)
- NMRC/OFLCG/003/2018/MOH | National Medical Research Council (NMRC)
- NRF-CRP19-2017-04/National Research Foundation Singapore (National Research Foundation-Prime Minister's office, Republic of Singapore)
LinkOut - more resources
Full Text Sources

