KAT8-mediated H4K16ac is essential for sustaining trophoblast self-renewal and proliferation via regulating CDX2
- PMID: 38961108
- PMCID: PMC11222414
- DOI: 10.1038/s41467-024-49930-6
KAT8-mediated H4K16ac is essential for sustaining trophoblast self-renewal and proliferation via regulating CDX2
Abstract
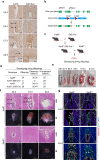
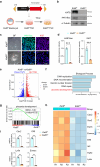
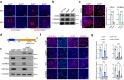
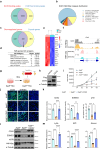
Abnormal trophoblast self-renewal and differentiation during early gestation is the major cause of miscarriage, yet the underlying regulatory mechanisms remain elusive. Here, we show that trophoblast specific deletion of Kat8, a MYST family histone acetyltransferase, leads to extraembryonic ectoderm abnormalities and embryonic lethality. Employing RNA-seq and CUT&Tag analyses on trophoblast stem cells (TSCs), we further discover that KAT8 regulates the transcriptional activation of the trophoblast stemness marker, CDX2, via acetylating H4K16. Remarkably, CDX2 overexpression partially rescues the defects arising from Kat8 knockout. Moreover, increasing H4K16ac via using deacetylase SIRT1 inhibitor, EX527, restores CDX2 levels and promoted placental development. Clinical analysis shows reduced KAT8, CDX2 and H4K16ac expression are associated with recurrent pregnancy loss (RPL). Trophoblast organoids derived from these patients exhibit impaired TSC self-renewal and growth, which are significantly ameliorated with EX527 treatment. These findings suggest the therapeutic potential of targeting the KAT8-H4K16ac-CDX2 axis for mitigating RPL, shedding light on early gestational abnormalities.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: An implication in early human pregnancy loss.Proc Natl Acad Sci U S A. 2020 Jul 28;117(30):17864-17875. doi: 10.1073/pnas.2002449117. Epub 2020 Jul 15. Proc Natl Acad Sci U S A. 2020. PMID: 32669432 Free PMC article.
-
MicroRNA regulation of murine trophoblast stem cell self-renewal and differentiation.Life Sci Alliance. 2020 Sep 9;3(11):e202000674. doi: 10.26508/lsa.202000674. Print 2020 Nov. Life Sci Alliance. 2020. PMID: 32907860 Free PMC article.
-
Deciphering a distinct regulatory network of TEAD4, CDX2 and GATA3 in humans for trophoblast transition from embryonic stem cells.Biochim Biophys Acta Mol Cell Res. 2020 Sep;1867(9):118736. doi: 10.1016/j.bbamcr.2020.118736. Epub 2020 May 7. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 32389642
-
Lysine Acetyltransferase 8: A Target for Natural Compounds in Cancer Therapy.Int J Mol Sci. 2025 May 29;26(11):5257. doi: 10.3390/ijms26115257. Int J Mol Sci. 2025. PMID: 40508066 Free PMC article. Review.
-
Progress in deciphering trophoblast cell differentiation during human placentation.Curr Opin Cell Biol. 2020 Dec;67:86-91. doi: 10.1016/j.ceb.2020.08.010. Epub 2020 Sep 18. Curr Opin Cell Biol. 2020. PMID: 32957014 Review.
Cited by
-
Gastrodin attenuates rheumatoid arthritis by targeting KAT8 to inhibit the lactylation of H3K9.Front Pharmacol. 2025 Mar 27;16:1559744. doi: 10.3389/fphar.2025.1559744. eCollection 2025. Front Pharmacol. 2025. PMID: 40213683 Free PMC article.
-
Abnormal lineage differentiation of peri-implantation aneuploid embryos revealed by single-cell RNA sequencing.Clin Transl Med. 2025 May;15(5):e70326. doi: 10.1002/ctm2.70326. Clin Transl Med. 2025. PMID: 40329849 Free PMC article. No abstract available.
-
Temporal and spatial distribution of histone acetylation in mouse molar development.PeerJ. 2025 Mar 31;13:e19215. doi: 10.7717/peerj.19215. eCollection 2025. PeerJ. 2025. PMID: 40183048 Free PMC article.
-
Elevated NGF provokes decidual lipid peroxidation and promotes preterm birth in mice.J Transl Med. 2025 Apr 28;23(1):481. doi: 10.1186/s12967-025-06424-3. J Transl Med. 2025. PMID: 40296138 Free PMC article.
-
Epigenetics of maternal-fetal interface immune microenvironment and placental related pregnancy complications.Front Immunol. 2025 Apr 3;16:1549839. doi: 10.3389/fimmu.2025.1549839. eCollection 2025. Front Immunol. 2025. PMID: 40248704 Free PMC article. Review.
References
-
- Cuffe, J. S., Holland, O., Salomon, C., Rice, G. E. & Perkins, A. V. J. P. Placental derived biomarkers of pregnancy disorders. Placenta54, 104–110 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- 82201861/National Natural Science Foundation of China (National Science Foundation of China)
- 82071652/National Natural Science Foundation of China (National Science Foundation of China)
- 82271695/National Natural Science Foundation of China (National Science Foundation of China)
- 82288102/National Natural Science Foundation of China (National Science Foundation of China)
- 2021M700945/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

