Deriving general structure-activity/selectivity relationship patterns for different subfamilies of cyclin-dependent kinase inhibitors using machine learning methods
- PMID: 38961127
- PMCID: PMC11222421
- DOI: 10.1038/s41598-024-66173-z
Deriving general structure-activity/selectivity relationship patterns for different subfamilies of cyclin-dependent kinase inhibitors using machine learning methods
Abstract
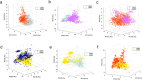
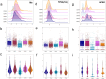
Cyclin-dependent kinases (CDKs) play essential roles in regulating the cell cycle and are among the most critical targets for cancer therapy and drug discovery. The primary objective of this research is to derive general structure-activity relationship (SAR) patterns for modeling the selectivity and activity levels of CDK inhibitors using machine learning methods. To accomplish this, 8592 small molecules with different binding affinities to CDK1, CDK2, CDK4, CDK5, and CDK9 were collected from Binding DB, and a diverse set of descriptors was calculated for each molecule. The supervised Kohonen networks (SKN) and counter propagation artificial neural networks (CPANN) models were trained to predict the activity levels and therapeutic targets of the molecules. The validity of models was confirmed through tenfold cross-validation and external test sets. Using selected sets of molecular descriptors (e.g. hydrophilicity and total polar surface area) we derived activity and selectivity maps to elucidate local regions in chemical space for active and selective CDK inhibitors. The SKN models exhibited prediction accuracies ranging from 0.75 to 0.94 for the external test sets. The developed multivariate classifiers were used for ligand-based virtual screening of 2 million random molecules of the PubChem database, yielding areas under the receiver operating characteristic curves ranging from 0.72 to 1.00 for the SKN model. Considering the persistent challenge of achieving CDK selectivity, this research significantly contributes to addressing the issue and underscores the paramount importance of developing drugs with minimized side effects.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
General structure-activity/selectivity relationship patterns for the inhibitors of the chemokine receptors (CCR1/CCR2/CCR4/CCR5) with application for virtual screening of PubChem database.J Biomol Struct Dyn. 2024 Oct;42(17):8781-8799. doi: 10.1080/07391102.2023.2248255. Epub 2023 Aug 20. J Biomol Struct Dyn. 2024. PMID: 37599469
-
Binding Activity Prediction of Cyclin-Dependent Inhibitors.J Chem Inf Model. 2015 Jul 27;55(7):1469-82. doi: 10.1021/ci500633c. Epub 2015 Jul 10. J Chem Inf Model. 2015. PMID: 26079845
-
Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases.Cancer Res. 2007 Sep 1;67(17):8325-34. doi: 10.1158/0008-5472.CAN-07-1826. Cancer Res. 2007. PMID: 17804748
-
Recent advances in the development of selective small molecule inhibitors for cyclin-dependent kinases.Curr Top Med Chem. 2005;5(2):167-79. doi: 10.2174/1568026053507688. Curr Top Med Chem. 2005. PMID: 15853645 Review.
-
Structural insights of cyclin dependent kinases: Implications in design of selective inhibitors.Eur J Med Chem. 2017 Dec 15;142:424-458. doi: 10.1016/j.ejmech.2017.08.071. Epub 2017 Sep 4. Eur J Med Chem. 2017. PMID: 28911822 Review.
Cited by
-
Computational design of CDK1 inhibitors with enhanced target affinity and drug-likeness using deep-learning framework.Heliyon. 2024 Nov 14;10(22):e40345. doi: 10.1016/j.heliyon.2024.e40345. eCollection 2024 Nov 30. Heliyon. 2024. PMID: 39748968 Free PMC article.
-
General structure-activity relationship models for the inhibitors of Adenosine receptors: A machine learning approach.Mol Divers. 2025 Aug;29(4):3253-3272. doi: 10.1007/s11030-024-11096-0. Epub 2025 Jan 20. Mol Divers. 2025. PMID: 39832081
References
-
- Alberts B, et al. Essential Cell Biology. Garland Science; 2015. pp. 603–610.
-
- Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of cdks, their cyclin activators, and cip and INK4 inhibitors1, 211998 Awardee, Walter J. Johnson Prize for the Encouragement of Research in the Life Sciences 2 Edited by P. E. Wright. J. Mol. Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

