Characterization, comparison, and phylogenetic analyses of chloroplast genomes of Euphorbia species
- PMID: 38961172
- PMCID: PMC11222452
- DOI: 10.1038/s41598-024-66102-0
Characterization, comparison, and phylogenetic analyses of chloroplast genomes of Euphorbia species
Abstract
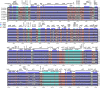
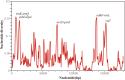
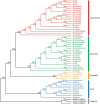
The genus Euphorbia (Euphorbiaceae) has near-cosmopolitan distribution and serves as a significant resource for both ornamental and medicinal purposes. Despite its economic importance, Euphorbia's taxonomy has long been challenged by the intricate nature of morphological traits exhibiting high levels of convergence. While molecular markers are essential for phylogenetic studies, their availability for Euphorbia has been limited. To address this gap, we conducted comparative analyses focusing on the chloroplast (CP) genomes of nine Euphorbia species, incorporating three newly sequenced and annotated accessions. In addition, phylogenetic informativeness and nucleotide diversity were computed to identify candidate markers for phylogenetic analyses among closely related taxa in the genus. Our investigation revealed relatively conserved sizes and structures of CP genomes across the studied species, with notable interspecific variations observed primarily in non-coding regions and IR/SC borders. By leveraging phylogenetic informativeness and nucleotide diversity, we identified rpoB gene as the optimal candidate for species delimitation and shallow-level phylogenetic inference within the genus. Through this comprehensive analysis of CP genomes across multiple taxa, our study sheds light on the evolutionary dynamics and taxonomic intricacies of Euphorbia, offering valuable insights into its CP genome evolution and taxonomy.
Keywords: Euphorbia; Chloroplast genome; Comparative analysis; Intraspecific variation; Phylogenetic analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Chloroplast genomics unravels taxonomic status and evolutionary relationships in Euphorbia fauriei and E. pekinensis.Gene. 2025 Feb 5;936:149116. doi: 10.1016/j.gene.2024.149116. Epub 2024 Nov 19. Gene. 2025. PMID: 39571662
-
Complete chloroplast genomes of eight Delphinium taxa (Ranunculaceae) endemic to Xinjiang, China: insights into genome structure, comparative analysis, and phylogenetic relationships.BMC Plant Biol. 2024 Jun 26;24(1):600. doi: 10.1186/s12870-024-05279-y. BMC Plant Biol. 2024. PMID: 38926811 Free PMC article.
-
Complete chloroplast genome of Stephania tetrandra (Menispermaceae) from Zhejiang Province: insights into molecular structures, comparative genome analysis, mutational hotspots and phylogenetic relationships.BMC Genomics. 2021 Dec 6;22(1):880. doi: 10.1186/s12864-021-08193-x. BMC Genomics. 2021. PMID: 34872502 Free PMC article.
-
Twelve newly assembled jasmine chloroplast genomes: unveiling genomic diversity, phylogenetic relationships and evolutionary patterns among Oleaceae and Jasminum species.BMC Plant Biol. 2024 Apr 25;24(1):331. doi: 10.1186/s12870-024-04995-9. BMC Plant Biol. 2024. PMID: 38664619 Free PMC article.
-
Exploring the phylogenetic framework and trait evolution of Impatiens through chloroplast genome analysis.BMC Plant Biol. 2024 Dec 20;24(1):1218. doi: 10.1186/s12870-024-05964-y. BMC Plant Biol. 2024. PMID: 39702025 Free PMC article.
Cited by
-
Intergeneric and interspecific relationships in tribe Ricineae revealed by phylogenomics of the plastome and transcriptome.Front Plant Sci. 2025 May 1;16:1544247. doi: 10.3389/fpls.2025.1544247. eCollection 2025. Front Plant Sci. 2025. PMID: 40376160 Free PMC article.
-
Phylogeny and evolutionary dynamics of the Rubia genus based on the chloroplast genome of Rubia tibetica.Sci Rep. 2025 Apr 24;15(1):14370. doi: 10.1038/s41598-025-96665-5. Sci Rep. 2025. PMID: 40274932 Free PMC article.
-
Untargeted Metabolomics and Targeted Phytohormone Profiling of Sweet Aloes (Euphorbia neriifolia) from Guyana: An Assessment of Asthma Therapy Potential in Leaf Extracts and Latex.Metabolites. 2025 Mar 5;15(3):177. doi: 10.3390/metabo15030177. Metabolites. 2025. PMID: 40137143 Free PMC article.
-
Comprehensive analysis of 385 chloroplast genomes unveils phylogenetic relationships and evolutionary history in cassava.BMC Plant Biol. 2025 Jul 3;25(1):858. doi: 10.1186/s12870-025-06883-2. BMC Plant Biol. 2025. PMID: 40610946 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

