Schlafen 11 further sensitizes BRCA-deficient cells to PARP inhibitors through single-strand DNA gap accumulation behind replication forks
- PMID: 38961202
- PMCID: PMC11315672
- DOI: 10.1038/s41388-024-03094-1
Schlafen 11 further sensitizes BRCA-deficient cells to PARP inhibitors through single-strand DNA gap accumulation behind replication forks
Abstract
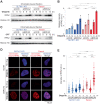
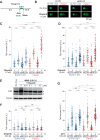
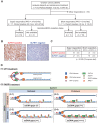
The preferential response to PARP inhibitors (PARPis) in BRCA-deficient and Schlafen 11 (SLFN11)-expressing ovarian cancers has been documented, yet the underlying molecular mechanisms remain unclear. As the accumulation of single-strand DNA (ssDNA) gaps behind replication forks is key for the lethality effect of PARPis, we investigated the combined effects of SLFN11 expression and BRCA deficiency on PARPi sensitivity and ssDNA gap formation in human cancer cells. PARPis increased chromatin-bound RPA2 and ssDNA gaps in SLFN11-expressing cells and even more in cells with BRCA1 or BRCA2 deficiency. SLFN11 was co-localized with chromatin-bound RPA2 under PARPis treatment, with enhanced recruitment in BRCA2-deficient cells. Notably, the chromatin-bound SLFN11 under PARPis did not block replication, contrary to its function under replication stress. SLFN11 recruitment was attenuated by the inactivation of MRE11. Hence, under PARPi treatment, MRE11 expression and BRCA deficiency lead to ssDNA gaps behind replication forks, where SLFN11 binds and increases their accumulation. As ovarian cancer patients who responded (progression-free survival >2 years) to olaparib maintenance therapy had a significantly higher SLFN11-positivity than short-responders (<6 months), our findings provide a mechanistic understanding of the favorable responses to PARPis in SLFN11-expressing and BRCA-deficient tumors. It highlight the clinical implications of SLFN11.
© 2024. The Author(s).
Conflict of interest statement
This work is supported by Ehime University PROS Grant for Intramural Collaboration (2022) (to J.M. and T.S.), and by the Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research (JP19H03505 and JP23H02768 to J.M.) and by the Japan Science and Technology Agency (JST) grant number JPMJFR2056 (JST FOREST Program to J.M). J.M. receives lecture fees from AstraZeneca plc and Takeda Pharmaceutical Company Limited. Y.K. receives lecture fees from AstraZeneca plc, Takeda Pharmaceutical Company Limited and ACTmed Company Limited. Other authors declare no potential competing interests.
Figures




Similar articles
-
Poly (ADP-Ribose) Polymerase Inhibitor Olaparib-Resistant BRCA1-Mutant Ovarian Cancer Cells Demonstrate Differential Sensitivity to PARP Inhibitor Rechallenge.Cells. 2024 Nov 7;13(22):1847. doi: 10.3390/cells13221847. Cells. 2024. PMID: 39594596 Free PMC article.
-
Reconsidering the mechanisms of action of PARP inhibitors based on clinical outcomes.Cancer Sci. 2022 Sep;113(9):2943-2951. doi: 10.1111/cas.15477. Epub 2022 Jul 16. Cancer Sci. 2022. PMID: 35766436 Free PMC article. Review.
-
PCAF-Mediated Histone Acetylation Promotes Replication Fork Degradation by MRE11 and EXO1 in BRCA-Deficient Cells.Mol Cell. 2020 Oct 15;80(2):327-344.e8. doi: 10.1016/j.molcel.2020.08.018. Epub 2020 Sep 22. Mol Cell. 2020. PMID: 32966758 Free PMC article.
-
Single-Stranded DNA Gap Accumulation Is a Functional Biomarker for USP1 Inhibitor Sensitivity.Cancer Res. 2024 Oct 15;84(20):3435-3446. doi: 10.1158/0008-5472.CAN-23-4007. Cancer Res. 2024. PMID: 38885312 Free PMC article.
-
Positioning loss of PARP1 activity as the central toxic event in BRCA-deficient cancer.DNA Repair (Amst). 2024 Dec;144:103775. doi: 10.1016/j.dnarep.2024.103775. Epub 2024 Oct 19. DNA Repair (Amst). 2024. PMID: 39461277 Review.
Cited by
-
From predictive biomarker to therapeutic target: the dual role of SLFN11 in chemotherapy sensitivity.Cancer Chemother Pharmacol. 2025 Jun 18;95(1):60. doi: 10.1007/s00280-025-04781-w. Cancer Chemother Pharmacol. 2025. PMID: 40531330 Review.
-
SLFN11, far from being limited to responding to cancer DNA damage.Clin Exp Med. 2025 Aug 26;25(1):304. doi: 10.1007/s10238-025-01776-y. Clin Exp Med. 2025. PMID: 40856812 Free PMC article. Review.
-
SLFN11-mediated ribosome biogenesis impairment induces TP53-independent apoptosis.Mol Cell. 2025 Mar 6;85(5):894-912.e10. doi: 10.1016/j.molcel.2025.01.008. Epub 2025 Feb 4. Mol Cell. 2025. PMID: 39909041
-
CAF-1 promotes efficient PrimPol recruitment to nascent DNA for single-stranded DNA gap formation.Nucleic Acids Res. 2024 Dec 11;52(22):13865-13880. doi: 10.1093/nar/gkae1068. Nucleic Acids Res. 2024. PMID: 39558157 Free PMC article.
-
Poly (ADP-ribose) polymerase inhibitors in cancer therapy.Chin Med J (Engl). 2025 Mar 20;138(6):634-650. doi: 10.1097/CM9.0000000000003471. Epub 2025 Feb 11. Chin Med J (Engl). 2025. PMID: 39932206 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

