Systematic characterization of immunoglobulin loci and deep sequencing of the expressed repertoire in the Atlantic cod (Gadus morhua)
- PMID: 38961347
- PMCID: PMC11223323
- DOI: 10.1186/s12864-024-10571-0
Systematic characterization of immunoglobulin loci and deep sequencing of the expressed repertoire in the Atlantic cod (Gadus morhua)
Abstract
Background: The Atlantic cod is a prolific species in the Atlantic, despite its inconsistent specific antibody response. It presents a peculiar case within vertebrate immunology due to its distinct immune system, characterized by the absence of MHCII antigen presentation pathway, required for T cell-dependent antibody responses. Thorough characterisation of immunoglobulin loci and analysis of the antibody repertoire is necessary to further our understanding of the Atlantic cod's immune response on a molecular level.
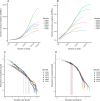
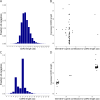
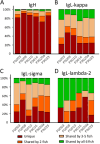
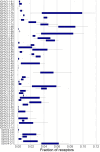
Results: A comprehensive search of the cod genome (gadmor3.0) identified the complete set of IgH genes organized into three sequential translocons on chromosome 2, while IgL genes were located on chromosomes 2 and 5. The Atlantic cod displayed a moderate germline V gene diversity, comprising four V gene families for both IgH and IgL, each with distinct chromosomal locations and organizational structures. 5'RACE sequencing revealed a diverse range of heavy chain CDR3 sequences and relatively limited CDR3 diversity in light chains. The analysis highlighted a differential impact of V-gene germline CDR3 length on receptor CDR3 length between heavy and light chains, underlining different recombination processes.
Conclusions: This study reveals that the Atlantic cod, despite its inconsistent antibody response, maintains a level of immunoglobulin diversity comparable to other fish species. The findings suggest that the extensive recent duplications of kappa light chain genes do not result in increased repertoire diversity. This research provides a comprehensive view of the Atlantic cod's immunoglobulin gene organization and repertoire, necessary for future studies of antibody responses at the molecular level.
Keywords: Atlantic cod; Diversity; Gadmor3.0; Ig; Immunogenetics; Repertoire.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Antibody repertoire development in teleosts--a review with emphasis on salmonids and Gadus morhua L.Dev Comp Immunol. 2006;30(1-2):57-76. doi: 10.1016/j.dci.2005.06.007. Dev Comp Immunol. 2006. PMID: 16084588 Review.
-
Identification of Igsigma and Iglambda in channel catfish, Ictalurus punctatus, and Iglambda in Atlantic cod, Gadus morhua.Immunogenetics. 2009 May;61(5):353-70. doi: 10.1007/s00251-009-0365-z. Epub 2009 Mar 31. Immunogenetics. 2009. PMID: 19333591
-
Characterization of extensive diversity in immunoglobulin light chain variable germline genes across biomedically important mouse strains.Immunohorizons. 2025 Jul 14;9(8):vlaf031. doi: 10.1093/immhor/vlaf031. Immunohorizons. 2025. PMID: 40682361 Free PMC article.
-
Ig-seq: Deep sequencing of the variable region of Atlantic salmon IgM heavy chain transcripts.Mol Immunol. 2017 Aug;88:99-105. doi: 10.1016/j.molimm.2017.06.022. Epub 2017 Jun 14. Mol Immunol. 2017. PMID: 28623734
-
The repertoire of human antibody to the Haemophilus influenzae type b capsular polysaccharide.Int Rev Immunol. 1992;9(1):25-43. doi: 10.3109/08830189209061781. Int Rev Immunol. 1992. PMID: 1484268 Review.
Cited by
-
Molecular characterization and complexity of the immunoglobulin repertoire in the silver-black fox (Vulpes vulpes).BMC Vet Res. 2025 Mar 28;21(1):214. doi: 10.1186/s12917-025-04676-1. BMC Vet Res. 2025. PMID: 40155920 Free PMC article.
-
Immunoglobulin gene loci structure and diversity of raccoon dog (Nyctereutes procyonoides).BMC Genomics. 2025 Apr 29;26(1):424. doi: 10.1186/s12864-025-11574-1. BMC Genomics. 2025. PMID: 40301716 Free PMC article.
-
Protective IgM-mediated immunity against Vibrio anguillarum in Atlantic cod with evolutionary losses of mhc class II and cd4.Front Immunol. 2025 Jul 2;16:1579541. doi: 10.3389/fimmu.2025.1579541. eCollection 2025. Front Immunol. 2025. PMID: 40672951 Free PMC article.
-
Exploring the Potential of Extracellular Vesicles from Atlantic Cod (Gadus morhua L.) Serum and Mucus for Wound Healing In Vitro.Biology (Basel). 2025 Jul 17;14(7):870. doi: 10.3390/biology14070870. Biology (Basel). 2025. PMID: 40723426 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

