Assembly and comparative analysis of the complete mitochondrial and chloroplast genome of Cyperus stoloniferus (Cyperaceae), a coastal plant possessing saline-alkali tolerance
- PMID: 38961375
- PMCID: PMC11220973
- DOI: 10.1186/s12870-024-05333-9
Assembly and comparative analysis of the complete mitochondrial and chloroplast genome of Cyperus stoloniferus (Cyperaceae), a coastal plant possessing saline-alkali tolerance
Abstract
Background: Cyperus stoloniferus is an important species in coastal ecosystems and possesses economic and ecological value. To elucidate the structural characteristics, variation, and evolution of the organelle genome of C. stoloniferus, we sequenced, assembled, and compared its mitochondrial and chloroplast genomes.
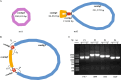
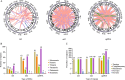
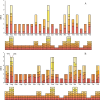
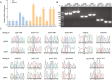
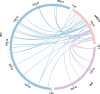
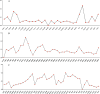
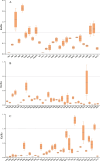
Results: We assembled the mitochondrial and chloroplast genomes of C. stoloniferus. The total length of the mitochondrial genome (mtDNA) was 927,413 bp, with a GC content of 40.59%. It consists of two circular DNAs, including 37 protein-coding genes (PCGs), 22 tRNAs, and five rRNAs. The length of the chloroplast genome (cpDNA) was 186,204 bp, containing 93 PCGs, 40 tRNAs, and 8 rRNAs. The mtDNA and cpDNA contained 81 and 129 tandem repeats, respectively, and 346 and 1,170 dispersed repeats, respectively, both of which have 270 simple sequence repeats. The third high-frequency codon (RSCU > 1) in the organellar genome tended to end at A or U, whereas the low-frequency codon (RSCU < 1) tended to end at G or C. The RNA editing sites of the PCGs were relatively few, with only 9 and 23 sites in the mtDNA and cpDNA, respectively. A total of 28 mitochondrial plastid DNAs (MTPTs) in the mtDNA were derived from cpDNA, including three complete trnT-GGU, trnH-GUG, and trnS-GCU. Phylogeny and collinearity indicated that the relationship between C. stoloniferus and C. rotundus are closest. The mitochondrial rns gene exhibited the greatest nucleotide variability, whereas the chloroplast gene with the greatest nucleotide variability was infA. Most PCGs in the organellar genome are negatively selected and highly evolutionarily conserved. Only six mitochondrial genes and two chloroplast genes exhibited Ka/Ks > 1; in particular, atp9, atp6, and rps7 may have undergone potential positive selection.
Conclusion: We assembled and validated the mtDNA of C. stoloniferus, which contains a 15,034 bp reverse complementary sequence. The organelle genome sequence of C. stoloniferus provides valuable genomic resources for species identification, evolution, and comparative genomic research in Cyperaceae.
Keywords: Cyperus stoloniferus; Comparative analysis; Systematic evolution; cpDNA; mtDNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Assembly and analysis of the complete mitochondrial genome of Leonurus japonicus (Lamiaceae).Sci Rep. 2025 Apr 18;15(1):13372. doi: 10.1038/s41598-025-97594-z. Sci Rep. 2025. PMID: 40246905 Free PMC article.
-
Mitochondrial genome assembly of the Chinese endemic species of Camellia luteoflora and revealing its repetitive sequence mediated recombination, codon preferences and MTPTs.BMC Plant Biol. 2025 Apr 5;25(1):435. doi: 10.1186/s12870-025-06461-6. BMC Plant Biol. 2025. PMID: 40186100 Free PMC article.
-
Complete Chloroplast Genome Sequence of the Endemic and Endangered Plant Dendropanax oligodontus: Genome Structure, Comparative and Phylogenetic Analysis.Genes (Basel). 2022 Nov 4;13(11):2028. doi: 10.3390/genes13112028. Genes (Basel). 2022. PMID: 36360265 Free PMC article.
-
Elucidating the multichromosomal structure within the Brasenia schreberi mitochondrial genome through assembly and analysis.BMC Genomics. 2024 Apr 29;25(1):422. doi: 10.1186/s12864-024-10331-0. BMC Genomics. 2024. PMID: 38684976 Free PMC article.
-
Organization and expression of organellar genomes.Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):785-97. doi: 10.1098/rstb.2009.0250. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124345 Free PMC article. Review.
Cited by
-
Characterize the Complete Mitogenome of Semiaquilegia guangxiensis and Assess the Efficiency of the Mitochondrial Genes in Ranunculales Phylogeny.Ecol Evol. 2025 Mar 27;15(4):e71165. doi: 10.1002/ece3.71165. eCollection 2025 Apr. Ecol Evol. 2025. PMID: 40170818 Free PMC article.
-
Chloroplast genome sequencing and phylogenetic analysis of Cyperus eragrostis lam. (cyperaceae).Mitochondrial DNA B Resour. 2025 Aug 8;10(9):795-799. doi: 10.1080/23802359.2025.2544680. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 40792195 Free PMC article.
-
Assembly and comparative analysis of the complete mitochondrial of Spodiopogon sagittifolius, an endemic and protective species from Yunnan, China.BMC Plant Biol. 2025 Mar 24;25(1):373. doi: 10.1186/s12870-025-06341-z. BMC Plant Biol. 2025. PMID: 40122803 Free PMC article.
-
Mitochondrial genome of Lonicera macranthoides: features, RNA editing, and insights into male sterility.Front Plant Sci. 2025 Jan 10;15:1520251. doi: 10.3389/fpls.2024.1520251. eCollection 2024. Front Plant Sci. 2025. PMID: 39866323 Free PMC article.
-
Characterization of the complete chloroplast genome and development of molecular markers of Salix.Sci Rep. 2024 Nov 18;14(1):28528. doi: 10.1038/s41598-024-79604-8. Sci Rep. 2024. PMID: 39557994 Free PMC article.
References
-
- Hayasaka D, Fujiwara K, Box EO. Recovery of sandy beach and maritime forest vegetation on Phuket island (Thailand) after the major Indian ocean tsunami of 2004. Appl Veg Sci. 2009;12(2):211–224. doi: 10.1111/j.1654-109X.2009.01017.x. - DOI
-
- Chau NM, Hong Hanh TT, Luyen NT, et al. Flavanones and stilbenes from Cyperus stoloniferus Retz. Biochem Syst Ecol. 2013;50:220–222. doi: 10.1016/j.bse.2013.04.004. - DOI
-
- Kumar, B. Cyperus Stoloniferus: The IUCN Red List of Threatened Species 2013: E.T177286A7406040,2010. 10.2305/IUCN.UK.2011-1.RLTS.T177286A7406040.en.
-
- Muasya AM, Reynders M, Goetghebeur P, et al. Dracoscirpoides (Cyperaceae) — a new genus from Southern Africa, its taxonomy and floral ontogeny. South Afr J Bot. 2012;78:104–115. doi: 10.1016/j.sajb.2011.05.011. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

