Integrating single-nucleus RNA sequencing and spatial transcriptomics to elucidate a specialized subpopulation of astrocytes, microglia and vascular cells in brains of mouse model of lipopolysaccharide-induced sepsis-associated encephalopathy
- PMID: 38961424
- PMCID: PMC11223438
- DOI: 10.1186/s12974-024-03161-0
Integrating single-nucleus RNA sequencing and spatial transcriptomics to elucidate a specialized subpopulation of astrocytes, microglia and vascular cells in brains of mouse model of lipopolysaccharide-induced sepsis-associated encephalopathy
Abstract
Background: Understanding the mechanism behind sepsis-associated encephalopathy (SAE) remains a formidable task. This study endeavors to shed light on the complex cellular and molecular alterations that occur in the brains of a mouse model with SAE, ultimately unraveling the underlying mechanisms of this condition.
Methods: We established a murine model using intraperitoneal injection of lipopolysaccharide (LPS) in wild type and Anxa1-/- mice and collected brain tissues for analysis at 0-hour, 12-hour, 24-hour, and 72-hour post-injection. Utilizing advanced techniques such as single-nucleus RNA sequencing (snRNA-seq) and Stereo-seq, we conducted a comprehensive characterization of the cellular responses and molecular patterns within the brain.
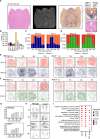
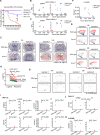
Results: Our study uncovered notable temporal differences in the response to LPS challenge between Anxa1-/- (annexin A1 knockout) and wild type mice, specifically at the 12-hour and 24-hour time points following injection. We observed a significant increase in the proportion of Astro-2 and Micro-2 cells in these mice. These cells exhibited a colocalization pattern with the vascular subtype Vas-1, forming a distinct region known as V1A2M2, where Astro-2 and Micro-2 cells surrounded Vas-1. Moreover, through further analysis, we discovered significant upregulation of ligands and receptors such as Timp1-Cd63, Timp1-Itgb1, Timp1-Lrp1, as well as Ccl2-Ackr1 and Cxcl2-Ackr1 within this region. In addition, we observed a notable increase in the expression of Cd14-Itgb1, Cd14-Tlr2, and Cd14-C3ar1 in regions enriched with Micro-2 cells. Additionally, Cxcl10-Sdc4 showed broad upregulation in brain regions containing both Micro-2 and Astro-2 cells. Notably, upon LPS challenge, there was an observed increase in Anxa1 expression in the mouse brain. Furthermore, our study revealed a noteworthy increase in mortality rates following Anxa1 knockdown. However, we did not observe substantial differences in the types, numbers, or distribution of other brain cells between Anxa1-/- and wildtype mice over time. Nevertheless, when comparing the 24-hour post LPS injection time point, we observed a significant decrease in the proportion and distribution of Micro-2 and Astro-2 cells in the vicinity of blood vessels in Anxa1-/- mice. Additionally, we noted reduced expression levels of several ligand-receptor pairs including Cd14-Tlr2, Cd14-C3ar1, Cd14-Itgb1, Cxcl10-Sdc4, Ccl2-Ackr1, and Cxcl2-Ackr1.
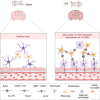
Conclusions: By combining snRNA-seq and Stereo-seq techniques, our study successfully identified a distinctive cellular colocalization, referred to as a special pathological niche, comprising Astro-2, Micro-2, and Vas-1 cells. Furthermore, we observed an upregulation of ligand-receptor pairs within this niche. These findings suggest a potential association between this cellular arrangement and the underlying mechanisms contributing to SAE or the increased mortality observed in Anxa1 knockdown mice.
Keywords: Anxa1; Cellular colocalization; Glial cell response; SAE; Sepsis; Single-nucleus RNA sequencing; Spatial transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Bulk and single-cell RNA-seq analyses reveal canonical RNA editing associated with microglia homeostasis and its role in sepsis-associated encephalopathy.Neuroscience. 2024 Nov 12;560:167-180. doi: 10.1016/j.neuroscience.2024.09.027. Epub 2024 Sep 16. Neuroscience. 2024. PMID: 39293730
-
TNFRSF6 induces mitochondrial dysfunction and microglia activation in the in vivo and in vitro models of sepsis-associated encephalopathy.Cell Mol Biol (Noisy-le-grand). 2024 Mar 31;70(3):102-109. doi: 10.14715/cmb/2024.70.3.15. Cell Mol Biol (Noisy-le-grand). 2024. PMID: 38650148
-
OTUD1 exacerbates sepsis-associated encephalopathy by promoting HK2 mitochondrial release to drive microglia pyroptosis.J Neuroinflammation. 2025 Jun 11;22(1):154. doi: 10.1186/s12974-025-03480-w. J Neuroinflammation. 2025. PMID: 40500776 Free PMC article.
-
Astroglia in Sepsis Associated Encephalopathy.Neurochem Res. 2020 Jan;45(1):83-99. doi: 10.1007/s11064-019-02743-2. Epub 2019 Feb 18. Neurochem Res. 2020. PMID: 30778837 Free PMC article. Review.
-
Sepsis-Associated Encephalopathy and Blood-Brain Barrier Dysfunction.Inflammation. 2021 Dec;44(6):2143-2150. doi: 10.1007/s10753-021-01501-3. Epub 2021 Jul 21. Inflammation. 2021. PMID: 34291398 Free PMC article. Review.
Cited by
-
A novel method for clustering cellular data to improve classification.Neural Regen Res. 2025 Sep 1;20(9):2697-2705. doi: 10.4103/NRR.NRR-D-24-00532. Epub 2024 Sep 24. Neural Regen Res. 2025. PMID: 39314166 Free PMC article.
-
Annexin A3 Represses Endothelial Permeability and Inflammation During Sepsis via Actin Cytoskeleton Modulation.Adv Sci (Weinh). 2025 Jun;12(22):e2416904. doi: 10.1002/advs.202416904. Epub 2025 Mar 28. Adv Sci (Weinh). 2025. PMID: 40151887 Free PMC article.
-
Sepsis-associated encephalopathy: Mechanisms, Diagnosis, and Treatments update.Int J Biol Sci. 2025 Apr 28;21(7):3214-3228. doi: 10.7150/ijbs.102234. eCollection 2025. Int J Biol Sci. 2025. PMID: 40384873 Free PMC article. Review.
-
Investigation of the Significance of Blood Signatures on Sepsis-Induced Acute Lung Injury in Sepsis Within 24 Hours.Int J Genomics. 2025 May 19;2025:5684300. doi: 10.1155/ijog/5684300. eCollection 2025. Int J Genomics. 2025. PMID: 40421173 Free PMC article.
-
LCN2 induces neuronal loss and facilitates sepsis-associated cognitive impairments.Cell Death Dis. 2025 Mar 1;16(1):146. doi: 10.1038/s41419-025-07469-4. Cell Death Dis. 2025. PMID: 40025014 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 2021A1515012429/Natural Science Foundation of Guangdong Province, China
- 211102114530659/Shaoguan Municipal Science and Technology Program, China
- 20221807/Shaoguan Engineering Research Center for Research and Development of Molecular and Cellular Technology in Rapid Diagnosis of Infectious Diseases and Cancer Program, China
- KEYANSHEN (2023) 01/Research Fund for Joint Laboratory for Digital and Precise Detection of Clinical Pathogens, Yuebei People's Hospital Affiliated to Shantou University Medical College, China
- RS202001/Research Project for Outstanding Scholar of Yuebei People's Hospital, Shantou University Medical College, China
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

