Antiviral activity of luteolin against porcine epidemic diarrhea virus in silico and in vitro
- PMID: 38961481
- PMCID: PMC11221151
- DOI: 10.1186/s12917-024-04053-4
Antiviral activity of luteolin against porcine epidemic diarrhea virus in silico and in vitro
Abstract
Background: Porcine epidemic diarrhea virus (PEDV) mainly causes acute and severe porcine epidemic diarrhea (PED), and is highly fatal in neonatal piglets. No reliable therapeutics against the infection exist, which poses a major global health issue for piglets. Luteolin is a flavonoid with anti-viral activity toward several viruses.
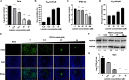
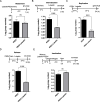
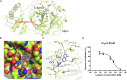
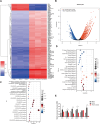
Results: We evaluated anti-viral effects of luteolin in PEDV-infected Vero and IPEC-J2 cells, and identified IC50 values of 23.87 µM and 68.5 µM, respectively. And found PEDV internalization, replication and release were significantly reduced upon luteolin treatment. As luteolin could bind to human ACE2 and SARS-CoV-2 main protease (Mpro) to contribute viral entry, we first identified that luteolin shares the same core binding site on pACE2 with PEDV-S by molecular docking and exhibited positive pACE2 binding with an affinity constant of 71.6 µM at dose-dependent increases by surface plasmon resonance (SPR) assay. However, pACE2 was incapable of binding to PEDV-S1. Therefore, luteolin inhibited PEDV internalization independent of PEDV-S binding to pACE2. Moreover, luteolin was firmly embedded in the groove of active pocket of Mpro in a three-dimensional docking model, and fluorescence resonance energy transfer (FRET) assays confirmed that luteolin inhibited PEDV Mpro activity. In addition, we also observed PEDV-induced pro-inflammatory cytokine inhibition and Nrf2-induced HO-1 expression. Finally, a drug resistant mutant was isolated after 10 cell culture passages concomitant with increasing luteolin concentrations, with reduced PEDV susceptibility to luteolin identified at passage 10.
Conclusions: Our results push forward that anti-PEDV mechanisms and resistant-PEDV properties for luteolin, which may be used to combat PED.
Keywords: Drug resistant mutant; Luteolin; Mpro; PEDV; Porcine ACE2; Pro-inflammatory cytokine; Spike.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Antiviral activity of flavonol against porcine epidemic diarrhea virus.Virology. 2024 Sep;597:110128. doi: 10.1016/j.virol.2024.110128. Epub 2024 Jun 1. Virology. 2024. PMID: 38861876
-
In silico and in vitro evaluation of antiviral activity of wogonin against main protease of porcine epidemic diarrhea virus.Front Cell Infect Microbiol. 2023 Mar 15;13:1123650. doi: 10.3389/fcimb.2023.1123650. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37009514 Free PMC article.
-
Ethyl caffeate as a novel targeted inhibitor of 3CLpro with antiviral activity against porcine epidemic diarrhea virus.Virology. 2025 Mar;604:110406. doi: 10.1016/j.virol.2025.110406. Epub 2025 Jan 16. Virology. 2025. PMID: 39854915
-
Coronavirus Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Interacts with p53 To Induce Cell Cycle Arrest in S-Phase and Promotes Viral Replication.J Virol. 2021 Jul 26;95(16):e0018721. doi: 10.1128/JVI.00187-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34037422 Free PMC article.
-
Aloe extract inhibits porcine epidemic diarrhea virus in vitro and in vivo.Vet Microbiol. 2020 Oct;249:108849. doi: 10.1016/j.vetmic.2020.108849. Epub 2020 Sep 15. Vet Microbiol. 2020. PMID: 32979750 Free PMC article.
Cited by
-
Luteolin alleviates Herpes Simplex Keratitis by inhibiting inflammatory responses via suppressing the PTGS2/NF-κB signaling pathway.Am J Transl Res. 2025 May 15;17(5):3307-3321. doi: 10.62347/IQUZ8416. eCollection 2025. Am J Transl Res. 2025. PMID: 40535652 Free PMC article.
-
Advances research in porcine enteric coronavirus therapies and antiviral drugs.Vet Q. 2024 Dec;44(1):1-49. doi: 10.1080/01652176.2024.2421299. Epub 2024 Nov 1. Vet Q. 2024. PMID: 39484691 Free PMC article. Review.
-
An overview of natural herbal extracts and compounds for combating porcine epidemic diarrhea virus.Front Vet Sci. 2025 Jun 6;12:1557198. doi: 10.3389/fvets.2025.1557198. eCollection 2025. Front Vet Sci. 2025. PMID: 40548251 Free PMC article. Review.
References
-
- Pathak RK, Kim WI, Kim JM. Targeting the PEDV 3CL protease for identification of small molecule inhibitors: an insight from virtual screening, ADMET prediction, molecular dynamics, free energy landscape, and binding energy calculations. J Biol Eng. 2023;17(1):29. doi: 10.1186/s13036-023-00342-y. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

