Making a Pathogen? Evaluating the Impact of Protist Predation on the Evolution of Virulence in Serratia marcescens
- PMID: 38961701
- PMCID: PMC11332436
- DOI: 10.1093/gbe/evae149
Making a Pathogen? Evaluating the Impact of Protist Predation on the Evolution of Virulence in Serratia marcescens
Abstract
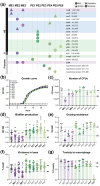
Opportunistic pathogens are environmental microbes that are generally harmless and only occasionally cause disease. Unlike obligate pathogens, the growth and survival of opportunistic pathogens do not rely on host infection or transmission. Their versatile lifestyles make it challenging to decipher how and why virulence has evolved in opportunistic pathogens. The coincidental evolution hypothesis postulates that virulence results from exaptation or pleiotropy, i.e. traits evolved for adaptation to living in one environment that have a different function in another. In particular, adaptation to avoid or survive protist predation has been suggested to contribute to the evolution of bacterial virulence (the training ground hypothesis). Here, we used experimental evolution to determine how the selective pressure imposed by a protist predator impacts the virulence and fitness of a ubiquitous environmental opportunistic bacterial pathogen that has acquired multidrug resistance: Serratia marcescens. To this aim, we evolved S. marcescens in the presence or absence of generalist protist predator, Tetrahymena thermophila. After 60 d of evolution, we evaluated genotypic and phenotypic changes by comparing evolved S. marcescens with the ancestral strain. Whole-genome shotgun sequencing of the entire evolved populations and individual isolates revealed numerous cases of parallel evolution, many more than statistically expected by chance, in genes associated with virulence. Our phenotypic assays suggested that evolution in the presence of a predator maintained virulence, whereas evolution in the absence of a predator resulted in attenuated virulence. We also found a significant correlation between virulence, biofilm formation, growth, and grazing resistance. Overall, our results provide evidence that bacterial virulence and virulence-related traits are maintained by selective pressures imposed by protist predation.
Keywords: bacteria; evolution; exaptation; opportunistic pathogen; predation; virulence.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest None declared.
Figures



Similar articles
-
Life history trade-offs and relaxed selection can decrease bacterial virulence in environmental reservoirs.PLoS One. 2012;7(8):e43801. doi: 10.1371/journal.pone.0043801. Epub 2012 Aug 24. PLoS One. 2012. PMID: 22937098 Free PMC article.
-
Predation on multiple trophic levels shapes the evolution of pathogen virulence.PLoS One. 2009 Aug 25;4(8):e6761. doi: 10.1371/journal.pone.0006761. PLoS One. 2009. PMID: 19707586 Free PMC article.
-
Within-host evolution decreases virulence in an opportunistic bacterial pathogen.BMC Evol Biol. 2015 Aug 19;15:165. doi: 10.1186/s12862-015-0447-5. BMC Evol Biol. 2015. PMID: 26282271 Free PMC article.
-
Dual Role of Mechanisms Involved in Resistance to Predation by Protozoa and Virulence to Humans.Front Microbiol. 2018 May 17;9:1017. doi: 10.3389/fmicb.2018.01017. eCollection 2018. Front Microbiol. 2018. PMID: 29867902 Free PMC article. Review.
-
Microbial warfare in the wild-the impact of protists on the evolution and virulence of bacterial pathogens.Int Microbiol. 2021 Nov;24(4):559-571. doi: 10.1007/s10123-021-00192-y. Epub 2021 Aug 8. Int Microbiol. 2021. PMID: 34365574 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

