Occurrence, distribution, and genetic diversity of faba bean viruses in China
- PMID: 38962134
- PMCID: PMC11219563
- DOI: 10.3389/fmicb.2024.1424699
Occurrence, distribution, and genetic diversity of faba bean viruses in China
Abstract
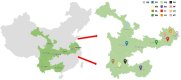
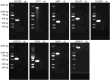
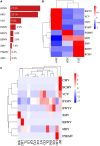
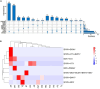
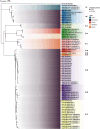
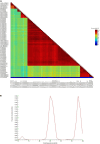
With worldwide cultivation, the faba bean (Vicia faba L.) stands as one of the most vital cool-season legume crops, serving as a major component of food security. China leads global faba bean production in terms of both total planting area and yield, with major production hubs in Yunnan, Sichuan, Jiangsu, and Gansu provinces. The faba bean viruses have caused serious yield losses in these production areas, but previous researches have not comprehensively investigated this issue. In this study, we collected 287 faba bean samples over three consecutive years from eight provinces/municipalities of China. We employed small RNA sequencing, RT-PCR, DNA sequencing, and phylogenetic analysis to detect the presence of viruses and examine their incidence, distribution, and genetic diversity. We identified a total of nine distinct viruses: bean yellow mosaic virus (BYMV, Potyvirus), milk vetch dwarf virus (MDV, Nanovirus), vicia cryptic virus (VCV, Alphapartitivirus), bean common mosaic virus (BCMV, Potyvirus), beet western yellows virus (BWYV, Polerovirus), broad bean wilt virus (BBWV, Fabavirus), soybean mosaic virus (SMV, Potyvirus), pea seed-borne mosaic virus (PSbMV, Potyvirus), and cucumber mosaic virus (CMV, Cucumovirus). BYMV was the predominant virus found during our sampling, followed by MDV and VCV. This study marks the first reported detection of BCMV in Chinese faba bean fields. Except for several isolates from Gansu and Yunnan provinces, our sequence analysis revealed that the majority of BYMV isolates contain highly conserved nucleotide sequences of coat protein (CP). Amino acid sequence alignment indicates that there is a conserved NAG motif at the N-terminal region of BYMV CP, which is considered important for aphid transmission. Our findings not only highlight the presence and diversity of pathogenic viruses in Chinese faba bean production, but also provide target pathogens for future antiviral resource screening and a basis for antiviral breeding.
Keywords: China; distribution; faba bean; genetic diversity; incidence; small RNA sequencing; viruses.
Copyright © 2024 Li, Qin, Zhu, Zhou, Zhao, Zhou, Wang, Chen and Cui.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bao S. Y., Wang X. M., Zhu Z. D., Zong X. X., Kumari S., Freeman A., et al. (2007). Survey of faba bean and field pea viruses in Yunnan Province, China. Australas. Plant Path. 36 347–353. 10.1071/AP07034 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

