ENT3C: an entropy-based similarity measure for Hi-C and micro-C derived contact matrices
- PMID: 38962256
- PMCID: PMC11217677
- DOI: 10.1093/nargab/lqae076
ENT3C: an entropy-based similarity measure for Hi-C and micro-C derived contact matrices
Abstract
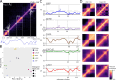
Hi-C and micro-C sequencing have shed light on the profound importance of 3D genome organization in cellular function by probing 3D contact frequencies across the linear genome. The resulting contact matrices are extremely sparse and susceptible to technical- and sequence-based biases, making their comparison challenging. The development of reliable, robust and efficient methods for quantifying similarity between contact matrices is crucial for investigating variations in the 3D genome organization in different cell types or under different conditions, as well as evaluating experimental reproducibility. We present a novel method, ENT3C, which measures the change in pattern complexity in the vicinity of contact matrix diagonals to quantify their similarity. ENT3C provides a robust, user-friendly Hi-C or micro-C contact matrix similarity metric and a characteristic entropy signal that can be used to gain detailed biological insights into 3D genome organization.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures
References
-
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002; 295:1306–1311. - PubMed
LinkOut - more resources
Full Text Sources

