Multicellular, IVT-derived, unmodified human transcriptome for nanopore-direct RNA analysis
- PMID: 38962390
- PMCID: PMC11221353
- DOI: 10.46471/gigabyte.129
Multicellular, IVT-derived, unmodified human transcriptome for nanopore-direct RNA analysis
Abstract
Nanopore direct RNA sequencing (DRS) enables measurements of RNA modifications. Modification-free transcripts are a practical and targeted control for DRS, providing a baseline measurement for canonical nucleotides within a matched and biologically-derived sequence context. However, these controls can be challenging to generate and carry nanopore-specific nuances that can impact analyses. We produced DRS datasets using modification-free transcripts from in vitro transcription of cDNA from six immortalized human cell lines. We characterized variation across cell lines and demonstrated how these may be interpreted. These data will serve as a versatile control and resource to the community for RNA modification analyses of human transcripts.
© The Author(s) 2024.
Conflict of interest statement
The authors declare no competing interests.
Figures
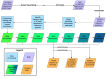
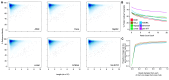
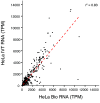
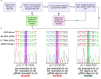
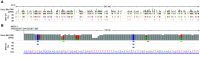
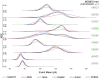
Update of
-
Multicellular, IVT-derived, unmodified human transcriptome for nanopore-direct RNA analysis.bioRxiv [Preprint]. 2024 May 28:2023.04.06.535889. doi: 10.1101/2023.04.06.535889. bioRxiv. 2024. Update in: GigaByte. 2024 Jun 17;2024:gigabyte129. doi: 10.46471/gigabyte.129. PMID: 37066160 Free PMC article. Updated. Preprint.
Similar articles
-
Multicellular, IVT-derived, unmodified human transcriptome for nanopore-direct RNA analysis.bioRxiv [Preprint]. 2024 May 28:2023.04.06.535889. doi: 10.1101/2023.04.06.535889. bioRxiv. 2024. Update in: GigaByte. 2024 Jun 17;2024:gigabyte129. doi: 10.46471/gigabyte.129. PMID: 37066160 Free PMC article. Updated. Preprint.
-
Transfer learning enables identification of multiple types of RNA modifications using nanopore direct RNA sequencing.Nat Commun. 2024 May 14;15(1):4049. doi: 10.1038/s41467-024-48437-4. Nat Commun. 2024. PMID: 38744925 Free PMC article.
-
mRNA psi profiling using nanopore DRS reveals cell type-specific pseudouridylation.bioRxiv [Preprint]. 2024 Nov 26:2024.05.08.593203. doi: 10.1101/2024.05.08.593203. bioRxiv. 2024. PMID: 38766185 Free PMC article. Preprint.
-
Computational methods for RNA modification detection from nanopore direct RNA sequencing data.RNA Biol. 2021 Oct 15;18(sup1):31-40. doi: 10.1080/15476286.2021.1978215. Epub 2021 Sep 24. RNA Biol. 2021. PMID: 34559589 Free PMC article. Review.
-
[Advances in mapping analysis of ribonucleic acid modifications through sequencing].Se Pu. 2024 Jul;42(7):632-645. doi: 10.3724/SP.J.1123.2023.12025. Se Pu. 2024. PMID: 38966972 Free PMC article. Review. Chinese.
Cited by
-
Direct RNA sequencing of the Escherichia coli epitranscriptome uncovers alterations under heat stress.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf175. doi: 10.1093/nar/gkaf175. Nucleic Acids Res. 2025. PMID: 40114376 Free PMC article.
-
Probing enzyme-dependent pseudouridylation using direct RNA sequencing to assess neuronal epitranscriptome plasticity.bioRxiv [Preprint]. 2024 Nov 26:2024.03.26.586895. doi: 10.1101/2024.03.26.586895. bioRxiv. 2024. Update in: Cell Syst. 2025 Apr 16;16(4):101238. doi: 10.1016/j.cels.2025.101238. PMID: 38585714 Free PMC article. Updated. Preprint.
-
Circuit logic: interdependent RNA modifications shape mRNA and noncoding RNA structure and function.RNA. 2025 Apr 16;31(5):613-622. doi: 10.1261/rna.080421.125. RNA. 2025. PMID: 40044218 Free PMC article. Review.
-
Prediction of m6A and m5C at single-molecule resolution reveals a transcriptome-wide co-occurrence of RNA modifications.Nat Commun. 2024 May 9;15(1):3899. doi: 10.1038/s41467-024-47953-7. Nat Commun. 2024. PMID: 38724548 Free PMC article.
-
Probing enzyme-dependent pseudouridylation using direct RNA sequencing to assess epitranscriptome plasticity in a neuronal cell line.Cell Syst. 2025 Apr 16;16(4):101238. doi: 10.1016/j.cels.2025.101238. Epub 2025 Mar 20. Cell Syst. 2025. PMID: 40118059
References
Grants and funding
LinkOut - more resources
Full Text Sources
